import plotly.express as px
import polars as pl
import polars.selectors as cs
import palmerpenguins
import nycflights1305 | Introduction to Data Science
Python data workflows with Polars and Plotly
1 Overview
In this beginning chapter, we will cover the fundamentals of data analysis in the form of importing, tidying, transforming, and visualizing data, as shown below:
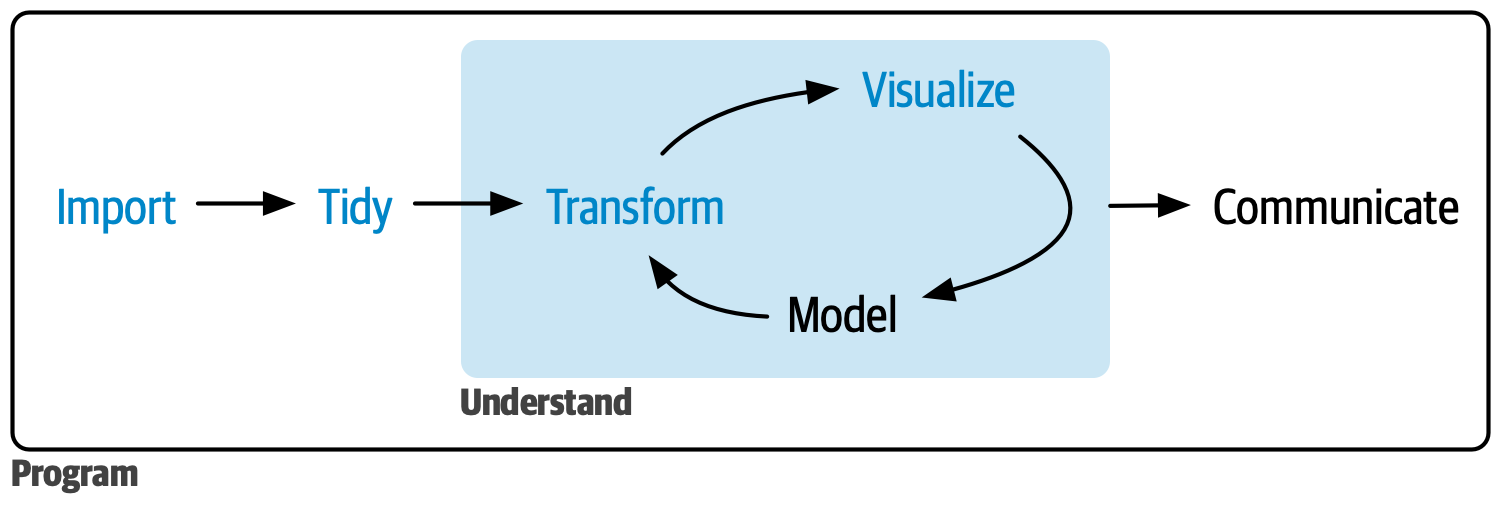
By the end of this module, you will be able to:
- Import data from various file formats into Python using Polars
- Clean and reshape datasets to facilitate analysis
- Transform data through filtering, sorting, and creating new variables
- Group and aggregate data to identify patterns and trends
- Visualize distributions and relationships using Plotly Express
- Apply tidy data principles to structure datasets effectively
1.1 Initialization
At this point, you will need to install a few more Python packages:
terminal
uv add polars[all] plotly[express] statsmodels palmerpenguins nycflights13Once installed, import the following:
2 Data visualization
Python has several systems for making graphs, but we will be focusing on Plotly, specifically Plotly Express. Plotly Express contains functions that can create entire plots at once, and makes it easy to create most common figures.
This section will teach you how to visualize your data using Plotly Express, walking through visualizing distributions of single variables, as well as relationships between two or more variables.
2.1 The
penguins data frame
The dataset we will be working with is a commonly used one,
affectionately referred to as the Palmer Penguins, which includes body
measurements for penguins on three islands in the Palmer Archipelago. A
data frame is a rectangular collection of variables (in
the columns) and observations (in the rows). penguins
contains 344 observations collected and made available by Dr. Kristen
Gorman and the Palmer Station, Antarctica.
Let’s define some term:
- A variable is a quantity, quality, or property that you can measure.
- A value is the state of a variable when you measure it. The value of a variable may change from measurement to measurement.
- An observation is a set of measurements made under similar conditions (you usually make all of the measurements in an observation at the same time and on the same object). An observation will contain several values, each associated with a different variable. We’ll sometimes refer to an observation as a data point.
- Tabular data is a set of values, each associated with a variable and an observation. Tabular data is tidy if each value is placed in its own “cell”, each variable in its own column, and each observation in its own row.
In this context, a variable refers to an attribute of all the penguins, and an observation refers to all the attributes of a single penguin.
We will use palmerpenguins package to get the
penguins data, and convert it to a polars data
frame:
penguins = pl.from_pandas(palmerpenguins.load_penguins())The reason we convert to a Polars data frame is because we want to use the tools and methods that come with Polars:
type(penguins)polars.dataframe.frame.DataFrameDepending on what tool/IDE you’re using Python with, just having the
variable name (penguins) as the last line will print a
formatted view of the data. If not, you can also use
print() to use Polars’ native formatting:
penguins| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | year |
|---|---|---|---|---|---|---|---|
| str | str | f64 | f64 | f64 | f64 | str | i64 |
| "Adelie" | "Torgersen" | 39.1 | 18.7 | 181.0 | 3750.0 | "male" | 2007 |
| "Adelie" | "Torgersen" | 39.5 | 17.4 | 186.0 | 3800.0 | "female" | 2007 |
| "Adelie" | "Torgersen" | 40.3 | 18.0 | 195.0 | 3250.0 | "female" | 2007 |
| "Adelie" | "Torgersen" | null | null | null | null | null | 2007 |
| … | … | … | … | … | … | … | … |
| "Chinstrap" | "Dream" | 43.5 | 18.1 | 202.0 | 3400.0 | "female" | 2009 |
| "Chinstrap" | "Dream" | 49.6 | 18.2 | 193.0 | 3775.0 | "male" | 2009 |
| "Chinstrap" | "Dream" | 50.8 | 19.0 | 210.0 | 4100.0 | "male" | 2009 |
| "Chinstrap" | "Dream" | 50.2 | 18.7 | 198.0 | 3775.0 | "female" | 2009 |
print(penguins)shape: (344, 8)
┌───────────┬───────────┬──────────────┬──────────────┬──────────────┬─────────────┬────────┬──────┐
│ species ┆ island ┆ bill_length_ ┆ bill_depth_m ┆ flipper_leng ┆ body_mass_g ┆ sex ┆ year │
│ --- ┆ --- ┆ mm ┆ m ┆ th_mm ┆ --- ┆ --- ┆ --- │
│ str ┆ str ┆ --- ┆ --- ┆ --- ┆ f64 ┆ str ┆ i64 │
│ ┆ ┆ f64 ┆ f64 ┆ f64 ┆ ┆ ┆ │
╞═══════════╪═══════════╪══════════════╪══════════════╪══════════════╪═════════════╪════════╪══════╡
│ Adelie ┆ Torgersen ┆ 39.1 ┆ 18.7 ┆ 181.0 ┆ 3750.0 ┆ male ┆ 2007 │
│ Adelie ┆ Torgersen ┆ 39.5 ┆ 17.4 ┆ 186.0 ┆ 3800.0 ┆ female ┆ 2007 │
│ Adelie ┆ Torgersen ┆ 40.3 ┆ 18.0 ┆ 195.0 ┆ 3250.0 ┆ female ┆ 2007 │
│ Adelie ┆ Torgersen ┆ null ┆ null ┆ null ┆ null ┆ null ┆ 2007 │
│ … ┆ … ┆ … ┆ … ┆ … ┆ … ┆ … ┆ … │
│ Chinstrap ┆ Dream ┆ 43.5 ┆ 18.1 ┆ 202.0 ┆ 3400.0 ┆ female ┆ 2009 │
│ Chinstrap ┆ Dream ┆ 49.6 ┆ 18.2 ┆ 193.0 ┆ 3775.0 ┆ male ┆ 2009 │
│ Chinstrap ┆ Dream ┆ 50.8 ┆ 19.0 ┆ 210.0 ┆ 4100.0 ┆ male ┆ 2009 │
│ Chinstrap ┆ Dream ┆ 50.2 ┆ 18.7 ┆ 198.0 ┆ 3775.0 ┆ female ┆ 2009 │
└───────────┴───────────┴──────────────┴──────────────┴──────────────┴─────────────┴────────┴──────┘This data frame contains 8 columns. For an alternative view, use
DataFrame.glimpse(), which is helpful for wide tables that
have many columns:
penguins.glimpse()Rows: 344
Columns: 8
$ species <str> 'Adelie', 'Adelie', 'Adelie', 'Adelie', 'Adelie', 'Adelie', 'Adelie', 'Adelie', 'Adelie', 'Adelie'
$ island <str> 'Torgersen', 'Torgersen', 'Torgersen', 'Torgersen', 'Torgersen', 'Torgersen', 'Torgersen', 'Torgersen', 'Torgersen', 'Torgersen'
$ bill_length_mm <f64> 39.1, 39.5, 40.3, None, 36.7, 39.3, 38.9, 39.2, 34.1, 42.0
$ bill_depth_mm <f64> 18.7, 17.4, 18.0, None, 19.3, 20.6, 17.8, 19.6, 18.1, 20.2
$ flipper_length_mm <f64> 181.0, 186.0, 195.0, None, 193.0, 190.0, 181.0, 195.0, 193.0, 190.0
$ body_mass_g <f64> 3750.0, 3800.0, 3250.0, None, 3450.0, 3650.0, 3625.0, 4675.0, 3475.0, 4250.0
$ sex <str> 'male', 'female', 'female', None, 'female', 'male', 'female', 'male', None, None
$ year <i64> 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007, 2007
Among these variables are:
species: a penguin’s species (Adelie, Chinstrap, or Gentoo).flipper_length_mm: length of a penguin’s flipper, in millimeters.body_mass_g: body mass of a penguin, in grams.
2.2 First visualization
Our goal is to recreate the following visual that displays the relationship between flipper lengths and body masses of these penguins, taking into consideration the species of the penguin.
2.3 Using Plotly Express
With Plotly Express, you begin a plot by calling a plotting function
from the module, commonly referred to as px. You can then
add arguments to your plot function for more customization.
At it’s most basic form, the plotting function creates a blank canvas with a grid since we’ve given it no data.
px.scatter()Next, we need to actually provide data, along with the appropriate number of variables depending on the type of plot we are trying to create.
px.scatter(data_frame=penguins, x="flipper_length_mm", y="body_mass_g")px.scatter() creates a scatter plot, and
we will learn many more plot types through out the course. You can learn
more about the different plots Plotly Express offers at their gallery.
This doesn’t match our “final goal” yet, but using this plot we can start answering the question that motivated our exploration: “What does the relationship between flipper length and body mass look like?” The relationship appears to be positive (as flipper length increases, so does body mass), fairly linear (the points are clustered around a line instead of a curve), and moderately strong (there isn’t too much scatter around such a line). Penguins with longer flippers are generally larger in terms of their body mass.
Before we go further, I want to point out that this dataset has some
missing values for flipper_length_mm and
body_mass_g, but Plotly does not warn you about this when
creating the plot. If one and/or other variable is missing data, we
cannot plot that.
2.4 Adding aesthetics and layers
Scatter plots are useful for displaying the relationship between two numerical variables, but it’s always a good idea to be skeptical of any apparent relationship between two variables and ask if there may be other variables that explain or change the nature of this apparent relationship. For example, does the relationship between flipper length and body mass differ by species? Let’s incorporate species into our plot and see if this reveals any additional insights into the apparent relationship between these variables. We will do this by representing species with different colored points.
To achieve this, we will use some of the other arguments that
px.scatter() provides for us, like color:
px.scatter(
data_frame=penguins,
x="flipper_length_mm",
y="body_mass_g",
color="species"
)When a categorical variable is mapped to an aesthetic, Plotly will automatically assign a unique value of the aesthetic (here, a unique color) to each unique level of the variable (each of the three species), a process known as scaling. Plotly will also add a legend that explains which value correspond to which levels.
Now let’s add another layer, a trendline displaying the relationship
between body mass and flipper length. px.scatter() has an
argument for this, trendline, and a couple other arguments
that modify its behavior. Specifically, we want to draw a line of best
fit using Ordinary Least Squares (ols). You can see the other options,
and more info about this plotting function with
?px.scatter() or in the online documentation.
px.scatter(
data_frame=penguins,
x="flipper_length_mm",
y="body_mass_g",
color="species",
trendline="ols"
)We’ve added lines, but this plot doesn’t look like our final goal,
which only has one line for the entire dataset, opposed to separate
lines for each of the penguin species. px.scatter() has an
argument, trendline_scope, which controls how the trendline
is drawn when there are groups, in this case created when we used
color="species". The default for
trendline_scope is "trace", which draws a line
per color, symbol, facet, etc., and "overall", which
computes one trendline for the entire dataset,a nd replicates across all
facets.
px.scatter(
data_frame=penguins,
x="flipper_length_mm",
y="body_mass_g",
color="species",
trendline="ols",
trendline_scope="overall"
)Now we have something that is very close to our final plot, thought it’s not there yet. We still need to use different shapes for each species and improve the labels.
It’s generally not a good idea to represent information only using
colors on a plot as people perceive colors differently do to color
blindness or other color vision difference. px.scatter()
allows us to control the shapes of the dots using the
symbol argument.
px.scatter(
penguins,
x="flipper_length_mm",
y="body_mass_g",
color="species",
symbol="species",
trendline="ols",
trendline_scope="overall",
)Note that the legend is automatically updated to reflect the different shapes of the points as well.
Finally, we can use title, subtitle, and
labels arguments to update our labels. title
and subtitle just take a string, adding labels are bit more
advanced. labels takes a dictionary with key:value combos
for each of the labels on the plot that you would like to change. In our
plot, we want to update the labels for the x-axis, y-axis, and the
legend, but we refer to them by their current label, not their
position:
px.scatter(
penguins,
x="flipper_length_mm",
y="body_mass_g",
color="species",
symbol="species",
trendline="ols",
trendline_scope="overall",
title="Body mass and flipper length",
subtitle="Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
labels={
"species":"Species",
"body_mass_g":"Body mass (g)",
"flipper_length_mm":"Flipper length (mm)"
}
)Now we have our final plot. If you haven’t noticed already, Plotly creates interactive plots, you can hover over certain data points to see their values, use the legend as a filter, and so on. In the top right of every plot, you will see a menu with some options.
2.5 Visualizing distributions
How you visualize the distribution of a variable depends on the type of the variable: categorical or numerical.
2.5.1 A categorical variable
A value is categorical if it can only take one of a
small set of values. To examine the distribution of a categorical
variable, you can use a bar chart. The height
of the bars displays how many observations occurred with each
x value.
px.bar(penguins, x="species")px.bar() will result in one rectangle drawn per
row of input, which can result in the striped look above. To
combine these rectangles into on color per position, we can
pre-calculate the count as and use it as the
y value:
# we'll learn more about this Polars code later
penguins_count = penguins.group_by("species").len("count")
print(penguins_count)
px.bar(penguins_count, x="species", y="count")shape: (3, 2)
┌───────────┬───────┐
│ species ┆ count │
│ --- ┆ --- │
│ str ┆ u32 │
╞═══════════╪═══════╡
│ Adelie ┆ 152 │
│ Gentoo ┆ 124 │
│ Chinstrap ┆ 68 │
└───────────┴───────┘The order of categorical values in axes, legends, and facets depends
on the order in which these values are first encountered in
data_frame. It’s often preferable to re-order the bars
based on their frequency, which we can do with the
category_orders argument. category_orders
takes a dictionary where the keys correspond to column names, and the
values should be lists of strings corresponding to the specific display
order desired
px.bar(
penguins_count,
x="species",
y="count",
category_orders={"species": ["Adelie", "Gentoo", "Chinstrap"]}
)While it’s easy enough to manually sort three columns, this could become very tedious for more columns. Here is one programmatic way you could sort the columns:
penguins_sorted = (
penguins_count
.sort(by="count", descending=True)
.get_column("species")
)
print(penguins_sorted)
px.bar(
penguins_count,
x="species",
y="count",
category_orders={"species": penguins_sorted}
)shape: (3,)
Series: 'species' [str]
[
"Adelie"
"Gentoo"
"Chinstrap"
]We will dive into the data manipulation code later, this is just to show what’s possible.
2.5.2 A numerical variable
A variable is numerical (or quantitative) if it can take on a wide range of numerical values, and it is sensible to add, subtract, or take averages with those values. Numerical variables can be continuous or discrete.
One common visualization for distributions of continuous variables is a histogram.
px.histogram(penguins, x="body_mass_g")A histogram divides the x-axis into equally spaced bins and then uses
the heigh of the bar to display the number observations that fall in
each bin. In the graph above, the tallest bar shows that 39 observations
have a body_mass_g value between 3,500 and 3,700 grams,
which are the left and right edges of the bar.
When working with histograms, it’s a good idea to use different number of bins to reveal different patterns in the data. In the plots below, X bars is too many, resulting in narrow bars. Similarly, 3 bins is too few, resulting in all the data being binned into huge categories that make it difficult to determine the shape of the distribution. A bin number of 20 provides a sensible balance.
px.histogram(penguins, x="body_mass_g", nbins=200)
px.histogram(penguins, x="body_mass_g", nbins=3)
px.histogram(penguins, x="body_mass_g", nbins=20)An alternative visualization for distributions of numerical variables is a density plot. A density plot is a smoothed-out version of a histogram and a practical alternative, particularly for continuous data that comes from an underlying smooth distribution. At the time of writing, Plotly Express doesn’t have a quick way to create a density plot, but it does offer very customizable violin plots, which we can make to look like a density plot if we would like.
px.violin(penguins, x="body_mass_g")The density plot is similar to the violin plot, with only one side, and the peaks are more exaggerated:
px.violin(
penguins,
x="body_mass_g", # plots the variable across the x-axis
range_y=[
0, # limits the bottom of y-axis, removing reflection
0.25 # limits the top of y-axis, stretching the peaks
]
)While this workaround works, sticking to the original violin plot I think looks better, and we can add some extra arguments to see more details:
px.violin(penguins, x="body_mass_g", points="all")Play around with different arguments and see what you like best!
As an analogy to understand these plots vs a histogram, imagine a histogram made out of wooden blocks. Then, imagine that you drop a cooked spaghetti string over it. The shape the spaghetti will take draped over blocks can be thought of as the shape of the density curve. It shows fewer details than a histogram but can make it easier to quickly glean the shape of the distribution, particularly with respect to modes and skewness.
2.6 Visualizing relationships
To visualize a relationship we need to have at least two variables mapped to aesthetics of a plot. In the following sections you will learn about commonly used plots for visualizing relationships between two or more variables and the plots used for creating them.
2.6.1 A numerical and a categorical variable
To visualize the relationship between a numerical and a categorical variable we can use side-by-side box plots. A boxplot is a type of visual shorthand for measures of position (percentiles) that describe a distribution. It is also useful for identifying potential outliers. Each boxplot consists of:
A box that indicates the range of the middle half of the data, a distance known as the inter-quartile range (IQR), stretching from the 25th percentile of the distribution to the 75th percentile. In the middle of the box is a line that displays the median, i.e. 50th percentile, of the distribution. These three lines give you a sense of the spread of the distribution and whether or not the distribution is symmetric about the median or skewed to one side.
Visual points that display observations that fall more than 1.5 times the IQR from either edge of the box. These outlying points are unusual so are plotted individually.
A line (or whisker) that extends from each end of the box and goes to the farthest non-outlier point in the distribution.
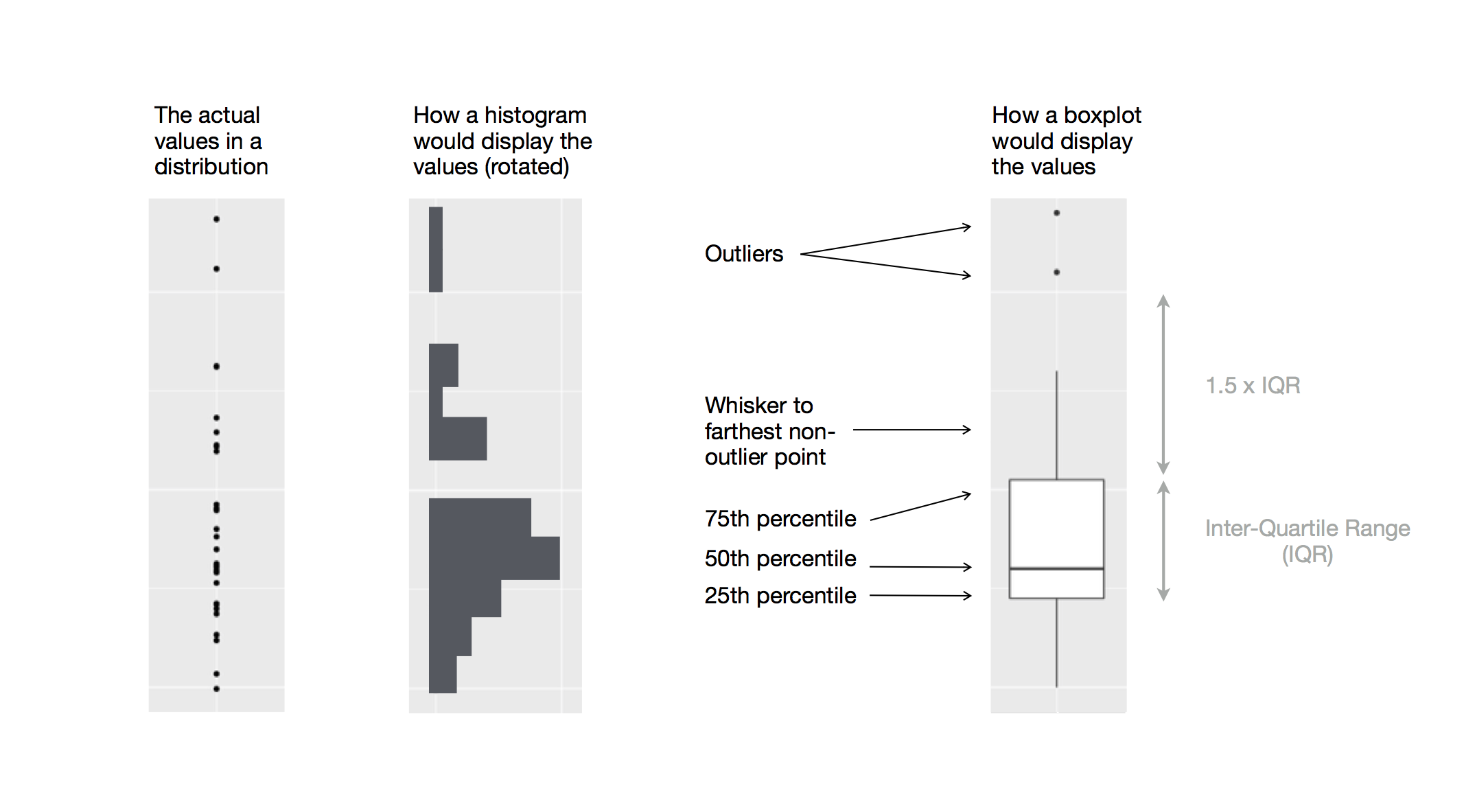
Let’s take a look at the distribution of body mass by species using
px.box()
px.box(penguins, x="species", y="body_mass_g")Alternatively, we can make violin plots with multiple groups:
px.violin(penguins, x="body_mass_g", color="species", box=True)As we’ve seen before, there are many ways to see and code what we are looking for.
2.6.2 Two categorical variables
We can use stacked bar plots to visualize the relationship between
two categorical variables. For example, the following two stacked bar
plots both display the relationship between island and
species, or specifically, visualizing the distribution of
species within each island.
The first plot shows the frequencies of each species of penguins on each island. The plot of frequencies shows that there are equal numbers of Adelies on each island. But we don’t have a good sense of the percentage balance within each island.
px.bar(penguins, x="island", color="species")or
data = penguins.group_by(["island", "species"]).len("count")
data_order = data.group_by("island").agg(pl.col("count").sum()).sort(by="count", descending=True).get_column("island")
px.bar(
data, x="island", y="count", color="species",
category_orders = {"island": data_order, "species": data_order}
)2.6.3 Two numerical variables
So far we’ve seen scatter plots for visualizing the relationship between two numerical variables. A scatter plot is probably the most commonly used plot for visualizing the relationship between to numerical variables.
px.scatter(penguins, x="flipper_length_mm", y="body_mass_g")2.6.4 Three or more variables
As we saw before, we can incorporate more variables into a plot by mapping them to additional aesthetics. For example, in the following plot, the colors of points represent species and the shapes represent islands.
px.scatter(
penguins,
x="flipper_length_mm", y="body_mass_g",
color="species", symbol="island"
)However adding too many aesthetic mappings to a plot makes it cluttered and difficult to make sense of. Another way, which is particularly useful for categorical variables, is to split your plot into facets, subplots that each display one subset of the data.
Most Plotly Express functions provide arguments to facet, just make sure to check the documentation.
px.scatter(
penguins,
x="flipper_length_mm", y="body_mass_g",
color="species", symbol="island",
facet_col="island"
)2.7 Summary
In this section, you’ve learned the basics of data visualization with Plotly Express. We started with the basic idea that underpins Plotly Express: a visualization is a mapping from variables in your data to aesthetic properties like position, color, size and shape. You then learned about increasing the complexity with more arguments in the Plotly functions. You also learned about commonly used plots for visualizing the distribution of a single variable as well as for visualizing relationships between two or more variables, by leveraging additional aesthetic mappings and/or splitting your plot into small multiples using faceting.
3 Data transformation
Visualization is an important tool for generating insight, but it’s rare that you get the data in exactly the right form you need to make the graph you want. Often you’ll need to create some new variables or summaries to answer your questions with your data, or maybe you just want to rename the variables or reorder the observations to make the data a little easier to work with. We’ve already seen examples of this above when creating the bar charts. In this section, we’ll see how to do that with the Polars package.
The goal of this section is to give you an overview of all the key tools for transforming a data frame. We’ll start with functions that operate on rows and then columns of a data frame, then circle back to talk more about method chaining, an important tool that you use to combine functions. We will then introduce the ability to work with groups. We will end the section with a case study that showcases these functions in action.
3.1 The
flights data frame
To explore basic Polars methods and expressions, we will use the
flights data frame from the nycflights13
package. This dataset contains all 336,776 flights that departed from
New York City in 2013.
flights = (
pl.from_pandas(nycflights13.flights)
.with_columns(pl.col("time_hour").str.to_datetime("%FT%TZ"))
) # We will learn what's going on here in later this section
flights| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | 517.0 | 515 | 2.0 | 830.0 | 819 | 11.0 | "UA" | 1545 | "N14228" | "EWR" | "IAH" | 227.0 | 1400 | 5 | 15 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 533.0 | 529 | 4.0 | 850.0 | 830 | 20.0 | "UA" | 1714 | "N24211" | "LGA" | "IAH" | 227.0 | 1416 | 5 | 29 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 542.0 | 540 | 2.0 | 923.0 | 850 | 33.0 | "AA" | 1141 | "N619AA" | "JFK" | "MIA" | 160.0 | 1089 | 5 | 40 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 544.0 | 545 | -1.0 | 1004.0 | 1022 | -18.0 | "B6" | 725 | "N804JB" | "JFK" | "BQN" | 183.0 | 1576 | 5 | 45 | 2013-01-01 10:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 9 | 30 | null | 2200 | null | null | 2312 | null | "9E" | 3525 | null | "LGA" | "SYR" | null | 198 | 22 | 0 | 2013-10-01 02:00:00 |
| 2013 | 9 | 30 | null | 1210 | null | null | 1330 | null | "MQ" | 3461 | "N535MQ" | "LGA" | "BNA" | null | 764 | 12 | 10 | 2013-09-30 16:00:00 |
| 2013 | 9 | 30 | null | 1159 | null | null | 1344 | null | "MQ" | 3572 | "N511MQ" | "LGA" | "CLE" | null | 419 | 11 | 59 | 2013-09-30 15:00:00 |
| 2013 | 9 | 30 | null | 840 | null | null | 1020 | null | "MQ" | 3531 | "N839MQ" | "LGA" | "RDU" | null | 431 | 8 | 40 | 2013-09-30 12:00:00 |
flights is a Polars DataFrame.
Different packages have their own version of a data frame with their own
methods, functions, etc., but in this course, we will be using Polars.
Polars provides its own way of working with data frames, as well as
importing, exporting, printing, and much more, including the previously
shown DataFrame.glimpse() method:
flights.glimpse()Rows: 336776
Columns: 19
$ year <i64> 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013, 2013
$ month <i64> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1
$ day <i64> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1
$ dep_time <f64> 517.0, 533.0, 542.0, 544.0, 554.0, 554.0, 555.0, 557.0, 557.0, 558.0
$ sched_dep_time <i64> 515, 529, 540, 545, 600, 558, 600, 600, 600, 600
$ dep_delay <f64> 2.0, 4.0, 2.0, -1.0, -6.0, -4.0, -5.0, -3.0, -3.0, -2.0
$ arr_time <f64> 830.0, 850.0, 923.0, 1004.0, 812.0, 740.0, 913.0, 709.0, 838.0, 753.0
$ sched_arr_time <i64> 819, 830, 850, 1022, 837, 728, 854, 723, 846, 745
$ arr_delay <f64> 11.0, 20.0, 33.0, -18.0, -25.0, 12.0, 19.0, -14.0, -8.0, 8.0
$ carrier <str> 'UA', 'UA', 'AA', 'B6', 'DL', 'UA', 'B6', 'EV', 'B6', 'AA'
$ flight <i64> 1545, 1714, 1141, 725, 461, 1696, 507, 5708, 79, 301
$ tailnum <str> 'N14228', 'N24211', 'N619AA', 'N804JB', 'N668DN', 'N39463', 'N516JB', 'N829AS', 'N593JB', 'N3ALAA'
$ origin <str> 'EWR', 'LGA', 'JFK', 'JFK', 'LGA', 'EWR', 'EWR', 'LGA', 'JFK', 'LGA'
$ dest <str> 'IAH', 'IAH', 'MIA', 'BQN', 'ATL', 'ORD', 'FLL', 'IAD', 'MCO', 'ORD'
$ air_time <f64> 227.0, 227.0, 160.0, 183.0, 116.0, 150.0, 158.0, 53.0, 140.0, 138.0
$ distance <i64> 1400, 1416, 1089, 1576, 762, 719, 1065, 229, 944, 733
$ hour <i64> 5, 5, 5, 5, 6, 5, 6, 6, 6, 6
$ minute <i64> 15, 29, 40, 45, 0, 58, 0, 0, 0, 0
$ time_hour <datetime[μs]> 2013-01-01 10:00:00, 2013-01-01 10:00:00, 2013-01-01 10:00:00, 2013-01-01 10:00:00, 2013-01-01 11:00:00, 2013-01-01 10:00:00, 2013-01-01 11:00:00, 2013-01-01 11:00:00, 2013-01-01 11:00:00, 2013-01-01 11:00:00
In both views, the variable names are followed by abbreviations that
tell you the type of each variable: i64 is short for
integer, f64 is short for float, str is short
for string, and datetime[μs] for date-time (in this case,
down to the micro-seconds).
We’re going to learn the primary methods (or contexts as Polars calls them) which will allow yo uto solve the vast majority of your data manipulation challenges. Before we discuss their individual differences, it’s worth stating what they have in common:
- The methods are always attached (or chained) to a data frame.
- The arguments typically describe which columns to operate on.
- The output is a new data frame (for the most part, ex:
group_by).
Because each method does one thing well, solving complex problems
will usually require combining multiple methods, and we will do so with
something called “method chaining”. You’ve already seen this before,
this is when we attach multiple methods together without creating a
placeholder variable between steps. You can think of each .
operator of saying “then”. This should help you get a sense of the
following code without understanding the details:
flights.filter(
pl.col("dest") == "IAH"
).group_by(
["year", "month", "day"]
).agg( # "aggregate", or summarize
arr_delay=pl.col("arr_delay").mean()
)| year | month | day | arr_delay |
|---|---|---|---|
| i64 | i64 | i64 | f64 |
| 2013 | 9 | 8 | -21.5 |
| 2013 | 5 | 27 | -25.095238 |
| 2013 | 11 | 29 | -25.384615 |
| 2013 | 7 | 17 | 14.7 |
| … | … | … | … |
| 2013 | 9 | 19 | 2.0 |
| 2013 | 5 | 1 | -2.473684 |
| 2013 | 4 | 9 | -1.526316 |
| 2013 | 3 | 3 | -36.65 |
We can also write the previous code in a cleaner format. When
surrounded by parenthesis, the . operator does not have to
“touch” the closing method before it:
(
flights
.filter(pl.col("dest") == "IAH")
.group_by(["year", "month", "day"])
.agg(arr_delay=pl.col("arr_delay").mean())
)If we didn’t use method chaining, we would have to create a bunch of intermediate objects:
flights1 = flights.filter(pl.col("dest") == "IAH")
flights2 = flights1.group_by(["year", "month", "day"])
flights3 = flights2.agg(arr_delay=pl.col("arr_delay").mean())While all of these have their time and place, method chaining generally produces data analysis code that is easier to write and read.
We can organize these contexts (methods) based on what they operate on: rows, columns, groups, or tables.
3.2 Rows
The most important contexts that operate on rows of a dataset are
DataFrame.filter(), which changes which rows are present
without changing their order, and DataFrame.sort(), which
changes the order of the rows without changing which are present. Both
methods only affect the rows, and the columns are left unchanged. We’ll
also see DataFrame.unique() which returns rows with unique
values. Unlike DataFrame.sort() and
DataFrame.filter(), it can also optionally modify the
columns.
3.2.1
DataFrame.filter()
DataFrame.filter()
allows you to keep rows based on the values of the columns. The
arguments (also known as predicates) are the conditions that must be
true to keep the row. For example, we could find all flights that
departed more than 120 minutes late:
flights.filter(pl.col("dep_delay") > 120)| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | 848.0 | 1835 | 853.0 | 1001.0 | 1950 | 851.0 | "MQ" | 3944 | "N942MQ" | "JFK" | "BWI" | 41.0 | 184 | 18 | 35 | 2013-01-01 23:00:00 |
| 2013 | 1 | 1 | 957.0 | 733 | 144.0 | 1056.0 | 853 | 123.0 | "UA" | 856 | "N534UA" | "EWR" | "BOS" | 37.0 | 200 | 7 | 33 | 2013-01-01 12:00:00 |
| 2013 | 1 | 1 | 1114.0 | 900 | 134.0 | 1447.0 | 1222 | 145.0 | "UA" | 1086 | "N76502" | "LGA" | "IAH" | 248.0 | 1416 | 9 | 0 | 2013-01-01 14:00:00 |
| 2013 | 1 | 1 | 1540.0 | 1338 | 122.0 | 2020.0 | 1825 | 115.0 | "B6" | 705 | "N570JB" | "JFK" | "SJU" | 193.0 | 1598 | 13 | 38 | 2013-01-01 18:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 9 | 30 | 1951.0 | 1649 | 182.0 | 2157.0 | 1903 | 174.0 | "EV" | 4294 | "N13988" | "EWR" | "SAV" | 95.0 | 708 | 16 | 49 | 2013-09-30 20:00:00 |
| 2013 | 9 | 30 | 2053.0 | 1815 | 158.0 | 2310.0 | 2054 | 136.0 | "EV" | 5292 | "N600QX" | "EWR" | "ATL" | 91.0 | 746 | 18 | 15 | 2013-09-30 22:00:00 |
| 2013 | 9 | 30 | 2159.0 | 1845 | 194.0 | 2344.0 | 2030 | 194.0 | "9E" | 3320 | "N906XJ" | "JFK" | "BUF" | 50.0 | 301 | 18 | 45 | 2013-09-30 22:00:00 |
| 2013 | 9 | 30 | 2235.0 | 2001 | 154.0 | 59.0 | 2249 | 130.0 | "B6" | 1083 | "N804JB" | "JFK" | "MCO" | 123.0 | 944 | 20 | 1 | 2013-10-01 00:00:00 |
We can use all of the same boolean expressions we’ve learned
previous, as well as chain them with & (instead of
and), | (instead of or), and
~ (instead of not). Note that Polars is picky
about ambiguity, so each condition we check for also has it’s own
parenthesis, similar to what we might use in a calculator to make sure
the order of operations is being followed exactly as we want:
# Flights that departed on January 1
flights.filter(
(pl.col("month") == 1) & (pl.col("day") == 1)
)| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | 517.0 | 515 | 2.0 | 830.0 | 819 | 11.0 | "UA" | 1545 | "N14228" | "EWR" | "IAH" | 227.0 | 1400 | 5 | 15 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 533.0 | 529 | 4.0 | 850.0 | 830 | 20.0 | "UA" | 1714 | "N24211" | "LGA" | "IAH" | 227.0 | 1416 | 5 | 29 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 542.0 | 540 | 2.0 | 923.0 | 850 | 33.0 | "AA" | 1141 | "N619AA" | "JFK" | "MIA" | 160.0 | 1089 | 5 | 40 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 544.0 | 545 | -1.0 | 1004.0 | 1022 | -18.0 | "B6" | 725 | "N804JB" | "JFK" | "BQN" | 183.0 | 1576 | 5 | 45 | 2013-01-01 10:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 1 | 1 | null | 1630 | null | null | 1815 | null | "EV" | 4308 | "N18120" | "EWR" | "RDU" | null | 416 | 16 | 30 | 2013-01-01 21:00:00 |
| 2013 | 1 | 1 | null | 1935 | null | null | 2240 | null | "AA" | 791 | "N3EHAA" | "LGA" | "DFW" | null | 1389 | 19 | 35 | 2013-01-02 00:00:00 |
| 2013 | 1 | 1 | null | 1500 | null | null | 1825 | null | "AA" | 1925 | "N3EVAA" | "LGA" | "MIA" | null | 1096 | 15 | 0 | 2013-01-01 20:00:00 |
| 2013 | 1 | 1 | null | 600 | null | null | 901 | null | "B6" | 125 | "N618JB" | "JFK" | "FLL" | null | 1069 | 6 | 0 | 2013-01-01 11:00:00 |
# Flights that departed in January or February
flights.filter(
(pl.col("month") == 1) | (pl.col("month") == 2)
)| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | 517.0 | 515 | 2.0 | 830.0 | 819 | 11.0 | "UA" | 1545 | "N14228" | "EWR" | "IAH" | 227.0 | 1400 | 5 | 15 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 533.0 | 529 | 4.0 | 850.0 | 830 | 20.0 | "UA" | 1714 | "N24211" | "LGA" | "IAH" | 227.0 | 1416 | 5 | 29 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 542.0 | 540 | 2.0 | 923.0 | 850 | 33.0 | "AA" | 1141 | "N619AA" | "JFK" | "MIA" | 160.0 | 1089 | 5 | 40 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 544.0 | 545 | -1.0 | 1004.0 | 1022 | -18.0 | "B6" | 725 | "N804JB" | "JFK" | "BQN" | 183.0 | 1576 | 5 | 45 | 2013-01-01 10:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 2 | 28 | null | 905 | null | null | 1115 | null | "MQ" | 4478 | "N722MQ" | "LGA" | "DTW" | null | 502 | 9 | 5 | 2013-02-28 14:00:00 |
| 2013 | 2 | 28 | null | 1115 | null | null | 1310 | null | "MQ" | 4485 | "N725MQ" | "LGA" | "CMH" | null | 479 | 11 | 15 | 2013-02-28 16:00:00 |
| 2013 | 2 | 28 | null | 830 | null | null | 1205 | null | "UA" | 1480 | null | "EWR" | "SFO" | null | 2565 | 8 | 30 | 2013-02-28 13:00:00 |
| 2013 | 2 | 28 | null | 840 | null | null | 1147 | null | "UA" | 443 | null | "JFK" | "LAX" | null | 2475 | 8 | 40 | 2013-02-28 13:00:00 |
There’s a useful shortcut when you’re combining | and
==: Expr.is_in(). It keeps rows where the
variable equals one of the values on the right:
# A shorter way to select flights that departed in January or February
flights.filter(
pl.col("month").is_in([1, 2])
)| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | 517.0 | 515 | 2.0 | 830.0 | 819 | 11.0 | "UA" | 1545 | "N14228" | "EWR" | "IAH" | 227.0 | 1400 | 5 | 15 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 533.0 | 529 | 4.0 | 850.0 | 830 | 20.0 | "UA" | 1714 | "N24211" | "LGA" | "IAH" | 227.0 | 1416 | 5 | 29 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 542.0 | 540 | 2.0 | 923.0 | 850 | 33.0 | "AA" | 1141 | "N619AA" | "JFK" | "MIA" | 160.0 | 1089 | 5 | 40 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 544.0 | 545 | -1.0 | 1004.0 | 1022 | -18.0 | "B6" | 725 | "N804JB" | "JFK" | "BQN" | 183.0 | 1576 | 5 | 45 | 2013-01-01 10:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 2 | 28 | null | 905 | null | null | 1115 | null | "MQ" | 4478 | "N722MQ" | "LGA" | "DTW" | null | 502 | 9 | 5 | 2013-02-28 14:00:00 |
| 2013 | 2 | 28 | null | 1115 | null | null | 1310 | null | "MQ" | 4485 | "N725MQ" | "LGA" | "CMH" | null | 479 | 11 | 15 | 2013-02-28 16:00:00 |
| 2013 | 2 | 28 | null | 830 | null | null | 1205 | null | "UA" | 1480 | null | "EWR" | "SFO" | null | 2565 | 8 | 30 | 2013-02-28 13:00:00 |
| 2013 | 2 | 28 | null | 840 | null | null | 1147 | null | "UA" | 443 | null | "JFK" | "LAX" | null | 2475 | 8 | 40 | 2013-02-28 13:00:00 |
When you run DataFrame.filter(), Polars executes the
filtering operation, creating a new DataFrame, and then prints it. It
doesn’t modify the existing flights dataset because Polars
never modifies the input (unless when explicitly chosen). To save the
result, you need to use the assignment operator, =:
jan1 = flights.filter((pl.col("month") == 1) & (pl.col("day") == 1))
jan1| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | 517.0 | 515 | 2.0 | 830.0 | 819 | 11.0 | "UA" | 1545 | "N14228" | "EWR" | "IAH" | 227.0 | 1400 | 5 | 15 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 533.0 | 529 | 4.0 | 850.0 | 830 | 20.0 | "UA" | 1714 | "N24211" | "LGA" | "IAH" | 227.0 | 1416 | 5 | 29 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 542.0 | 540 | 2.0 | 923.0 | 850 | 33.0 | "AA" | 1141 | "N619AA" | "JFK" | "MIA" | 160.0 | 1089 | 5 | 40 | 2013-01-01 10:00:00 |
| 2013 | 1 | 1 | 544.0 | 545 | -1.0 | 1004.0 | 1022 | -18.0 | "B6" | 725 | "N804JB" | "JFK" | "BQN" | 183.0 | 1576 | 5 | 45 | 2013-01-01 10:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 1 | 1 | null | 1630 | null | null | 1815 | null | "EV" | 4308 | "N18120" | "EWR" | "RDU" | null | 416 | 16 | 30 | 2013-01-01 21:00:00 |
| 2013 | 1 | 1 | null | 1935 | null | null | 2240 | null | "AA" | 791 | "N3EHAA" | "LGA" | "DFW" | null | 1389 | 19 | 35 | 2013-01-02 00:00:00 |
| 2013 | 1 | 1 | null | 1500 | null | null | 1825 | null | "AA" | 1925 | "N3EVAA" | "LGA" | "MIA" | null | 1096 | 15 | 0 | 2013-01-01 20:00:00 |
| 2013 | 1 | 1 | null | 600 | null | null | 901 | null | "B6" | 125 | "N618JB" | "JFK" | "FLL" | null | 1069 | 6 | 0 | 2013-01-01 11:00:00 |
3.2.2
DataFrame.sort()
DataFrame.sort()
changes the order of the rows based on the value of the columns. It
takes a data frame and a set of column names (or more complicated
expressions) to order by. If you provide more than one column name, each
additional column will be used to break ties in the values of the
preceding columns. For example, the following code sorts by the
departure time, which is spread over four columns. We get the earliest
years first, then within a year, the earliest months, etc.
flights.sort(by=["year", "month", "day", "dep_time"])| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | null | 1935 | null | null | 2240 | null | "AA" | 791 | "N3EHAA" | "LGA" | "DFW" | null | 1389 | 19 | 35 | 2013-01-02 00:00:00 |
| 2013 | 1 | 1 | null | 1500 | null | null | 1825 | null | "AA" | 1925 | "N3EVAA" | "LGA" | "MIA" | null | 1096 | 15 | 0 | 2013-01-01 20:00:00 |
| 2013 | 1 | 1 | null | 1630 | null | null | 1815 | null | "EV" | 4308 | "N18120" | "EWR" | "RDU" | null | 416 | 16 | 30 | 2013-01-01 21:00:00 |
| 2013 | 1 | 1 | null | 600 | null | null | 901 | null | "B6" | 125 | "N618JB" | "JFK" | "FLL" | null | 1069 | 6 | 0 | 2013-01-01 11:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 12 | 31 | 2328.0 | 2330 | -2.0 | 412.0 | 409 | 3.0 | "B6" | 1389 | "N651JB" | "EWR" | "SJU" | 198.0 | 1608 | 23 | 30 | 2014-01-01 04:00:00 |
| 2013 | 12 | 31 | 2332.0 | 2245 | 47.0 | 58.0 | 3 | 55.0 | "B6" | 486 | "N334JB" | "JFK" | "ROC" | 60.0 | 264 | 22 | 45 | 2014-01-01 03:00:00 |
| 2013 | 12 | 31 | 2355.0 | 2359 | -4.0 | 430.0 | 440 | -10.0 | "B6" | 1503 | "N509JB" | "JFK" | "SJU" | 195.0 | 1598 | 23 | 59 | 2014-01-01 04:00:00 |
| 2013 | 12 | 31 | 2356.0 | 2359 | -3.0 | 436.0 | 445 | -9.0 | "B6" | 745 | "N665JB" | "JFK" | "PSE" | 200.0 | 1617 | 23 | 59 | 2014-01-01 04:00:00 |
You can use positional arguments to sort by multiple columns in the same way:
flights.sort(
by=["year", "month", "day", "dep_time"],
descending=[False, False, False, True]
)| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 1 | null | 600 | null | null | 901 | null | "B6" | 125 | "N618JB" | "JFK" | "FLL" | null | 1069 | 6 | 0 | 2013-01-01 11:00:00 |
| 2013 | 1 | 1 | null | 1500 | null | null | 1825 | null | "AA" | 1925 | "N3EVAA" | "LGA" | "MIA" | null | 1096 | 15 | 0 | 2013-01-01 20:00:00 |
| 2013 | 1 | 1 | null | 1935 | null | null | 2240 | null | "AA" | 791 | "N3EHAA" | "LGA" | "DFW" | null | 1389 | 19 | 35 | 2013-01-02 00:00:00 |
| 2013 | 1 | 1 | null | 1630 | null | null | 1815 | null | "EV" | 4308 | "N18120" | "EWR" | "RDU" | null | 416 | 16 | 30 | 2013-01-01 21:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 12 | 31 | 459.0 | 500 | -1.0 | 655.0 | 651 | 4.0 | "US" | 1895 | "N557UW" | "EWR" | "CLT" | 95.0 | 529 | 5 | 0 | 2013-12-31 10:00:00 |
| 2013 | 12 | 31 | 26.0 | 2245 | 101.0 | 129.0 | 2353 | 96.0 | "B6" | 108 | "N374JB" | "JFK" | "PWM" | 50.0 | 273 | 22 | 45 | 2014-01-01 03:00:00 |
| 2013 | 12 | 31 | 18.0 | 2359 | 19.0 | 449.0 | 444 | 5.0 | "DL" | 412 | "N713TW" | "JFK" | "SJU" | 192.0 | 1598 | 23 | 59 | 2014-01-01 04:00:00 |
| 2013 | 12 | 31 | 13.0 | 2359 | 14.0 | 439.0 | 437 | 2.0 | "B6" | 839 | "N566JB" | "JFK" | "BQN" | 189.0 | 1576 | 23 | 59 | 2014-01-01 04:00:00 |
Note that the number of rows has not changed, we’re only arranging the data, we’re not filtering it.
3.2.3
DataFrame.unique()
DataFrame.unique()
finds all the unique rows in a dataset, so technically, it primarily
operates on the rows. Most of the time, however, you’ll want the
distinct combination of some variables, so you can also optionally
supply column names:
# Remove duplicate rows, if any
flights.unique()| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 7 | 15 | 2107.0 | 2030 | 37.0 | 2213.0 | 2202 | 11.0 | "9E" | 4079 | "N8588D" | "JFK" | "BWI" | 37.0 | 184 | 20 | 30 | 2013-07-16 00:00:00 |
| 2013 | 2 | 12 | 1441.0 | 1445 | -4.0 | 1728.0 | 1744 | -16.0 | "B6" | 153 | "N558JB" | "JFK" | "MCO" | 146.0 | 944 | 14 | 45 | 2013-02-12 19:00:00 |
| 2013 | 6 | 16 | 809.0 | 815 | -6.0 | 1038.0 | 1033 | 5.0 | "DL" | 914 | "N351NW" | "LGA" | "DEN" | 234.0 | 1620 | 8 | 15 | 2013-06-16 12:00:00 |
| 2013 | 8 | 6 | 1344.0 | 1349 | -5.0 | 1506.0 | 1515 | -9.0 | "UA" | 643 | "N807UA" | "EWR" | "ORD" | 122.0 | 719 | 13 | 49 | 2013-08-06 17:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 5 | 26 | 1146.0 | 1130 | 16.0 | 1428.0 | 1428 | 0.0 | "B6" | 61 | "N653JB" | "JFK" | "FLL" | 146.0 | 1069 | 11 | 30 | 2013-05-26 15:00:00 |
| 2013 | 12 | 1 | 1656.0 | 1700 | -4.0 | 1757.0 | 1816 | -19.0 | "B6" | 1734 | "N283JB" | "JFK" | "BTV" | 47.0 | 266 | 17 | 0 | 2013-12-01 22:00:00 |
| 2013 | 6 | 28 | 700.0 | 700 | 0.0 | 953.0 | 1006 | -13.0 | "DL" | 1415 | "N662DN" | "JFK" | "SLC" | 263.0 | 1990 | 7 | 0 | 2013-06-28 11:00:00 |
| 2013 | 7 | 24 | 1057.0 | 1100 | -3.0 | 1338.0 | 1349 | -11.0 | "DL" | 695 | "N928DL" | "JFK" | "MCO" | 135.0 | 944 | 11 | 0 | 2013-07-24 15:00:00 |
# Find all unique origin and destination pairs
flights.unique(subset=["origin", "dest"])| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 1 | 2 | 1552.0 | 1600 | -8.0 | 1727.0 | 1725 | 2.0 | "EV" | 4922 | "N371CA" | "LGA" | "ROC" | 47.0 | 254 | 16 | 0 | 2013-01-02 21:00:00 |
| 2013 | 1 | 1 | 1832.0 | 1835 | -3.0 | 2059.0 | 2103 | -4.0 | "9E" | 3830 | "N8894A" | "JFK" | "CHS" | 106.0 | 636 | 18 | 35 | 2013-01-01 23:00:00 |
| 2013 | 1 | 1 | 1558.0 | 1534 | 24.0 | 1808.0 | 1703 | 65.0 | "EV" | 4502 | "N16546" | "EWR" | "BNA" | 168.0 | 748 | 15 | 34 | 2013-01-01 20:00:00 |
| 2013 | 1 | 1 | 810.0 | 810 | 0.0 | 1048.0 | 1037 | 11.0 | "9E" | 3538 | "N915XJ" | "JFK" | "MSP" | 189.0 | 1029 | 8 | 10 | 2013-01-01 13:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 1 | 1 | 805.0 | 805 | 0.0 | 1015.0 | 1005 | 10.0 | "B6" | 219 | "N273JB" | "JFK" | "CLT" | 98.0 | 541 | 8 | 5 | 2013-01-01 13:00:00 |
| 2013 | 1 | 1 | 1318.0 | 1322 | -4.0 | 1358.0 | 1416 | -18.0 | "EV" | 4106 | "N19554" | "EWR" | "BDL" | 25.0 | 116 | 13 | 22 | 2013-01-01 18:00:00 |
| 2013 | 1 | 2 | 905.0 | 822 | 43.0 | 1313.0 | 1045 | null | "EV" | 4140 | "N15912" | "EWR" | "XNA" | null | 1131 | 8 | 22 | 2013-01-02 13:00:00 |
| 2013 | 1 | 1 | 1840.0 | 1845 | -5.0 | 2055.0 | 2030 | 25.0 | "MQ" | 4517 | "N725MQ" | "LGA" | "CRW" | 96.0 | 444 | 18 | 45 | 2013-01-01 23:00:00 |
It should be noted that the default argument for keep is
any, which does not give any guarantee of which
unique rows are kept. If you dataset is ordered, you might want
to use one of the other options for keep.
If you want the number of occurrences instead, you’ll need to use a
combination of DataFrame.group_by along with a
GroupBy.agg().
3.3 Columns
There are two important contexts that affect the columns without
changing the rows: DataFrame.with_columns(), which creates
new columns that are derived from the existing columns, &
select(), which changes which columns are present.
3.3.1
DataFrame.with_columns()
The job of DataFrame.with_columns()
is to add new columns. In the Data Wrangling module, you will learn a
large set of functions that you can use to manipulate different types of
variables. For new, we’ll stick with basic algebra, which allows us to
compute the gain, how much time a delayed flight made up in
the air, and the speed in miles per hour:
flights.with_columns(
gain = pl.col("dep_delay") - pl.col("arr_delay"),
speed = pl.col("distance") / pl.col("air_time") * 60
)| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour | gain | speed |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] | f64 | f64 |
| 2013 | 1 | 1 | 517.0 | 515 | 2.0 | 830.0 | 819 | 11.0 | "UA" | 1545 | "N14228" | "EWR" | "IAH" | 227.0 | 1400 | 5 | 15 | 2013-01-01 10:00:00 | -9.0 | 370.044053 |
| 2013 | 1 | 1 | 533.0 | 529 | 4.0 | 850.0 | 830 | 20.0 | "UA" | 1714 | "N24211" | "LGA" | "IAH" | 227.0 | 1416 | 5 | 29 | 2013-01-01 10:00:00 | -16.0 | 374.273128 |
| 2013 | 1 | 1 | 542.0 | 540 | 2.0 | 923.0 | 850 | 33.0 | "AA" | 1141 | "N619AA" | "JFK" | "MIA" | 160.0 | 1089 | 5 | 40 | 2013-01-01 10:00:00 | -31.0 | 408.375 |
| 2013 | 1 | 1 | 544.0 | 545 | -1.0 | 1004.0 | 1022 | -18.0 | "B6" | 725 | "N804JB" | "JFK" | "BQN" | 183.0 | 1576 | 5 | 45 | 2013-01-01 10:00:00 | 17.0 | 516.721311 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 9 | 30 | null | 2200 | null | null | 2312 | null | "9E" | 3525 | null | "LGA" | "SYR" | null | 198 | 22 | 0 | 2013-10-01 02:00:00 | null | null |
| 2013 | 9 | 30 | null | 1210 | null | null | 1330 | null | "MQ" | 3461 | "N535MQ" | "LGA" | "BNA" | null | 764 | 12 | 10 | 2013-09-30 16:00:00 | null | null |
| 2013 | 9 | 30 | null | 1159 | null | null | 1344 | null | "MQ" | 3572 | "N511MQ" | "LGA" | "CLE" | null | 419 | 11 | 59 | 2013-09-30 15:00:00 | null | null |
| 2013 | 9 | 30 | null | 840 | null | null | 1020 | null | "MQ" | 3531 | "N839MQ" | "LGA" | "RDU" | null | 431 | 8 | 40 | 2013-09-30 12:00:00 | null | null |
Note that since we haven’t assigned the result of the above
computation back to flights, the new variables
gain and speed will only be printed but will
not be stored in a data frame. And if we want them to be available in a
data frame for future use, we should think carefully about whether we
want the result to be assigned back to flights, overwriting
the original data frame with many more variables, or to a new object.
Often, the right answer is a new object that is named informatively to
indicate its contents, e.g., delay_gain, but you might also
have good reasons for overwriting flights.
3.3.2
DataFrame.select()
It’s not uncommon to get datasets with hundreds or even thousands of
variables. In this situation, the first challenge is often just focusing
on the variables you’re interested in. DataFrame.select()
allows you to rapidly zoom in on a useful subset using operations based
on the names of the variables:
- Select columns by name:
flights.select("year")| year |
|---|
| i64 |
| 2013 |
| 2013 |
| 2013 |
| 2013 |
| … |
| 2013 |
| 2013 |
| 2013 |
| 2013 |
- Select multiple columns by passing a list of column names:
flights.select(["year", "month", "day"])| year | month | day |
|---|---|---|
| i64 | i64 | i64 |
| 2013 | 1 | 1 |
| 2013 | 1 | 1 |
| 2013 | 1 | 1 |
| 2013 | 1 | 1 |
| … | … | … |
| 2013 | 9 | 30 |
| 2013 | 9 | 30 |
| 2013 | 9 | 30 |
| 2013 | 9 | 30 |
- Multiple columns can also be selected using positional arguments instead of a list. Expressions are also accepted:
flights.select(
pl.col("year"),
pl.col("month"),
month_add_one = pl.col("month") + 1 # Adds 1 to the values of "month"
)| year | month | month_add_one |
|---|---|---|
| i64 | i64 | i64 |
| 2013 | 1 | 2 |
| 2013 | 1 | 2 |
| 2013 | 1 | 2 |
| 2013 | 1 | 2 |
| … | … | … |
| 2013 | 9 | 10 |
| 2013 | 9 | 10 |
| 2013 | 9 | 10 |
| 2013 | 9 | 10 |
Polars also provides more advanced way to select columns using its Selectors.
Selectors allow for more intuitive selection of columns from DataFrame
objects based on their name, type, or other properties. They unify and
build on the related functionality that is available through the
pl.col() expression and can also broadcast expressions over
the selected columns.
Selectors are available as functions imported from
polars.selectors. Typical/recommended usage is to import
the module as cs and employ selectors from there.
import polars as pl
import polars.selectors as csThere are a number of selectors you can use within select:
cs.starts_with("abc"): matches column names that begin with “abc”.cs.ends_with("xyz"): matches column names that end with “xyz”.cs.contains("ijk"): matches column names that contain “ijk”.cs.matches(r"\d{3}"): matches column names using regex, columns with three digits repeating in the name in this case.cs.temporal(): matches columns with temporal (time) data types.cs.string(): matches columns with string data types.
These are just a few, you can see all the selectors with examples in
the documentation, or by looking at the options after typing
cs. in your editor.
You can combine selectors with the following set
operations:
| Operation | Expression |
|---|---|
| UNION | A | B |
| INTERSECTION | A & B |
| DIFFERENCE | A - B |
| SYMMETRIC DIFFERENCE | A ^ B |
| COMPLEMENT | ~A |
flights.select(cs.temporal() | cs.string())| carrier | tailnum | origin | dest | time_hour |
|---|---|---|---|---|
| str | str | str | str | datetime[μs] |
| "UA" | "N14228" | "EWR" | "IAH" | 2013-01-01 10:00:00 |
| "UA" | "N24211" | "LGA" | "IAH" | 2013-01-01 10:00:00 |
| "AA" | "N619AA" | "JFK" | "MIA" | 2013-01-01 10:00:00 |
| "B6" | "N804JB" | "JFK" | "BQN" | 2013-01-01 10:00:00 |
| … | … | … | … | … |
| "9E" | null | "LGA" | "SYR" | 2013-10-01 02:00:00 |
| "MQ" | "N535MQ" | "LGA" | "BNA" | 2013-09-30 16:00:00 |
| "MQ" | "N511MQ" | "LGA" | "CLE" | 2013-09-30 15:00:00 |
| "MQ" | "N839MQ" | "LGA" | "RDU" | 2013-09-30 12:00:00 |
Note that both individual selector results and selector set operations will always return matching columns in the same order as the underlying DataFrame schema.
3.4 Groups
So far, we’ve learned about contexts (DataFrame methods) that work
with rows and columns. Polars gets even more powerful when
you add in the ability to work with groups. In this section, we’ll focus
on the most important contexts: DataFrame.group_by(),
DataFrame.agg(), and the various slice-ing
contexts.
3.4.1
DataFrame.group_by()
Use DataFrame.group_by()
to divide your dataset into groups meaningful for your analysis:
flights.group_by("month")<polars.dataframe.group_by.GroupBy at 0x1f439f85250>As you can see, DataFrame.group_by() doesn’t change the
data, but returns a GroupBy
object. This acts like a DataFrame, but subsequent
operations will now work “by month”, and comes with some extra
methods.
3.4.2
GroupBy.agg()
The most important grouped operation is an aggregation, which, if
being used to calculate a single summary statistic, reduces the data
frame to have a single row for each group. In Polars, this operation is
performed by GroupBy.agg(),
as shown by the following example, which computes the average departure
delay by month:
(
flights
.group_by("month")
.agg(avg_delay = pl.col("dep_delay").mean())
)| month | avg_delay |
|---|---|
| i64 | f64 |
| 1 | 10.036665 |
| 8 | 12.61104 |
| 12 | 16.576688 |
| 11 | 5.435362 |
| … | … |
| 6 | 20.846332 |
| 5 | 12.986859 |
| 3 | 13.227076 |
| 7 | 21.727787 |
There are two things to note:
- Polars automatically drops the missing values in
dep_delaywhen calculating the mean. - Polars has a few different methods called
mean(), but they do different things depending on root object (DataFrame,GroupBy,Expr, etc.)
You can create any number of aggregations in a single call to
GroupBy.agg(). You’ll learn various useful summaries in
later modules, but one very useful summary is pl.len(),
which returns the number of rows in each group:
(
flights
.group_by("month")
.agg(
avg_delay = pl.col("dep_delay").mean(),
n = pl.len()
)
.sort("month")
)| month | avg_delay | n |
|---|---|---|
| i64 | f64 | u32 |
| 1 | 10.036665 | 27004 |
| 2 | 10.816843 | 24951 |
| 3 | 13.227076 | 28834 |
| 4 | 13.938038 | 28330 |
| … | … | … |
| 9 | 6.722476 | 27574 |
| 10 | 6.243988 | 28889 |
| 11 | 5.435362 | 27268 |
| 12 | 16.576688 | 28135 |
Means and counts can get you a surprisingly long way in data science!
3.4.3 Slicing functions
There are a few ways Polars provides for you to extract specific rows within each group:
GroupBy.head(n = 1)takes the first row from each group. (Also works withDataFrame)GroupBy.tail(n = 1)takes the last row from each group. (Also works withDataFrame)
GroupBy also provides some powerful aggregations for
whole groups, like:
flights.group_by("dest").max() # shows max value for each group and column
flights.group_by("dest").min() # shows min value for each group and column| dest | year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| str | i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| "BUR" | 2013 | 1 | 1 | 1325.0 | 1329 | -11.0 | 6.0 | 1640 | -61.0 | "B6" | 355 | "N503JB" | "JFK" | 293.0 | 2465 | 13 | 0 | 2013-01-01 18:00:00 |
| "SEA" | 2013 | 1 | 1 | 39.0 | 638 | -21.0 | 2.0 | 17 | -75.0 | "AA" | 5 | "N11206" | "EWR" | 275.0 | 2402 | 6 | 0 | 2013-01-01 12:00:00 |
| "DAY" | 2013 | 1 | 1 | 41.0 | 739 | -19.0 | 3.0 | 933 | -59.0 | "9E" | 3805 | "N10156" | "EWR" | 71.0 | 533 | 7 | 0 | 2013-01-01 17:00:00 |
| "LAX" | 2013 | 1 | 1 | 2.0 | 551 | -16.0 | 1.0 | 3 | -75.0 | "AA" | 1 | "N11206" | "EWR" | 275.0 | 2454 | 5 | 0 | 2013-01-01 11:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| "ILM" | 2013 | 9 | 1 | 925.0 | 935 | -19.0 | 4.0 | 1143 | -49.0 | "EV" | 4885 | "N371CA" | "LGA" | 63.0 | 500 | 9 | 0 | 2013-09-07 13:00:00 |
| "TVC" | 2013 | 6 | 2 | 722.0 | 730 | -20.0 | 2.0 | 943 | -39.0 | "EV" | 3402 | "N11107" | "EWR" | 84.0 | 644 | 7 | 0 | 2013-06-14 22:00:00 |
| "OKC" | 2013 | 1 | 1 | 20.0 | 1630 | -13.0 | 3.0 | 1912 | -49.0 | "EV" | 4141 | "N10156" | "EWR" | 162.0 | 1325 | 16 | 0 | 2013-01-02 00:00:00 |
| "MKE" | 2013 | 1 | 1 | 14.0 | 559 | -20.0 | 1.0 | 720 | -63.0 | "9E" | 46 | "N10156" | "EWR" | 93.0 | 725 | 5 | 0 | 2013-01-01 12:00:00 |
If you want the top/bottom k number of rows (optionally
by group), use the DataFrame.top_k or
DataFrame.bottom_k contexts:
flights.top_k(k = 4, by = "arr_delay")
flights.bottom_k(k = 4, by = "arr_delay")| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 5 | 7 | 1715.0 | 1729 | -14.0 | 1944.0 | 2110 | -86.0 | "VX" | 193 | "N843VA" | "EWR" | "SFO" | 315.0 | 2565 | 17 | 29 | 2013-05-07 21:00:00 |
| 2013 | 5 | 20 | 719.0 | 735 | -16.0 | 951.0 | 1110 | -79.0 | "VX" | 11 | "N840VA" | "JFK" | "SFO" | 316.0 | 2586 | 7 | 35 | 2013-05-20 11:00:00 |
| 2013 | 5 | 2 | 1947.0 | 1949 | -2.0 | 2209.0 | 2324 | -75.0 | "UA" | 612 | "N851UA" | "EWR" | "LAX" | 300.0 | 2454 | 19 | 49 | 2013-05-02 23:00:00 |
| 2013 | 5 | 6 | 1826.0 | 1830 | -4.0 | 2045.0 | 2200 | -75.0 | "AA" | 269 | "N3KCAA" | "JFK" | "SEA" | 289.0 | 2422 | 18 | 30 | 2013-05-06 22:00:00 |
3.4.4 Grouping by multiple variables
You can create groups using more than one variable. For example, we could make a group for each date:
daily = flights.group_by(["year", "month", "day"])
daily.max()| year | month | day | dep_time | sched_dep_time | dep_delay | arr_time | sched_arr_time | arr_delay | carrier | flight | tailnum | origin | dest | air_time | distance | hour | minute | time_hour |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| i64 | i64 | i64 | f64 | i64 | f64 | f64 | i64 | f64 | str | i64 | str | str | str | f64 | i64 | i64 | i64 | datetime[μs] |
| 2013 | 9 | 22 | 2350.0 | 2359 | 239.0 | 2400.0 | 2359 | 232.0 | "YV" | 6181 | "N9EAMQ" | "LGA" | "XNA" | 626.0 | 4983 | 23 | 59 | 2013-09-23 03:00:00 |
| 2013 | 6 | 29 | 2342.0 | 2359 | 313.0 | 2358.0 | 2359 | 284.0 | "WN" | 6054 | "N999DN" | "LGA" | "TYS" | 605.0 | 4983 | 23 | 59 | 2013-06-30 03:00:00 |
| 2013 | 7 | 21 | 2356.0 | 2359 | 580.0 | 2400.0 | 2359 | 645.0 | "YV" | 6120 | "N9EAMQ" | "LGA" | "XNA" | 629.0 | 4983 | 23 | 59 | 2013-07-22 03:00:00 |
| 2013 | 9 | 4 | 2258.0 | 2359 | 296.0 | 2359.0 | 2359 | 148.0 | "YV" | 6101 | "N995AT" | "LGA" | "XNA" | 597.0 | 4983 | 23 | 59 | 2013-09-05 03:00:00 |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| 2013 | 5 | 9 | 2356.0 | 2359 | 504.0 | 2359.0 | 2359 | 493.0 | "YV" | 5769 | "N986DL" | "LGA" | "XNA" | 620.0 | 4983 | 23 | 59 | 2013-05-10 03:00:00 |
| 2013 | 2 | 13 | 2353.0 | 2359 | 592.0 | 2358.0 | 2359 | 595.0 | "YV" | 6055 | "N995AT" | "LGA" | "XNA" | 616.0 | 4983 | 23 | 59 | 2013-02-14 04:00:00 |
| 2013 | 2 | 28 | 2359.0 | 2359 | 168.0 | 2400.0 | 2358 | 145.0 | "YV" | 5739 | "N9EAMQ" | "LGA" | "XNA" | 612.0 | 4983 | 23 | 59 | 2013-03-01 04:00:00 |
| 2013 | 6 | 10 | 2352.0 | 2359 | 401.0 | 2359.0 | 2359 | 354.0 | "YV" | 6177 | "N9EAMQ" | "LGA" | "XNA" | 604.0 | 4983 | 23 | 59 | 2013-06-11 03:00:00 |
GroupBy methods return a DataFrame, so
there is no need to explicitly “un-group” your dataset.
3.5 Summary
In this chapter, you’ve learned the tools that Polars provides for
working with data frames. The tools are roughly grouped into three
categories: those that manipulate the rows (like
DataFrame.filter() and DataFrame.sort()) those
that manipulate the columns (like DataFrame.select() and
DataFrame.with_columns()) and those that manipulate groups
(like DataFrame.group_by() and GroupBy.agg()).
In this chapter, we’ve focused on these “whole data frame” tools, but
you haven’t yet learned much about what you can do with the individual
variable. We’ll return to that in a later module in the course, where
each section provides tools for a specific type of variable.
4 Data tidying
In this section, you will learn a consistent way to organize your data in Python using a system called tidy data. Getting your data into this format requires some work up front, but that work pays off in the long term. Once you have tidy data, you will spend much less time munging data from one representation to another, allowing you to spend more time on the data questions you care about.
You’ll first learn the definition of tidy data and see it applied to a simple toy dataset. Then we’ll dive into the primary tool you’ll use for tidying data: pivoting. Pivoting allows you to change the form of your data without changing any of the values.
4.1 Tidy data
You can represent the same underlying data in multiple ways. The
example below shows the same data organized in three different ways.
Each dataset shows the same values of four variables:
country, year, population, and
number of documented cases of TB (tuberculosis), but each
dataset organizes the values in a different way.
table1| country | year | cases | population |
|---|---|---|---|
| str | i64 | i64 | i64 |
| "Afghanistan" | 1999 | 745 | 19987071 |
| "Afghanistan" | 2000 | 2666 | 20595360 |
| "Brazil" | 1999 | 37737 | 172006362 |
| "Brazil" | 2000 | 80488 | 174504898 |
| "China" | 1999 | 212258 | 1272915272 |
| "China" | 2000 | 213766 | 1280428583 |
table2| country | year | type | count |
|---|---|---|---|
| str | i64 | str | i64 |
| "Afghanistan" | 1999 | "cases" | 745 |
| "Afghanistan" | 1999 | "population" | 19987071 |
| "Afghanistan" | 2000 | "cases" | 2666 |
| "Afghanistan" | 2000 | "population" | 20595360 |
| … | … | … | … |
| "China" | 1999 | "cases" | 212258 |
| "China" | 1999 | "population" | 1272915272 |
| "China" | 2000 | "cases" | 213766 |
| "China" | 2000 | "population" | 1280428583 |
table3| country | year | rate |
|---|---|---|
| str | i64 | str |
| "Afghanistan" | 1999 | "745/19987071" |
| "Afghanistan" | 2000 | "2666/20595360" |
| "Brazil" | 1999 | "37737/172006362" |
| "Brazil" | 2000 | "80488/174504898" |
| "China" | 1999 | "212258/1272915272" |
| "China" | 2000 | "213766/1280428583" |
These are all representations of the same underlying data, but they
are not equally easy to use. One of them, table1, will be
much easier to work with Polars & Plotly because it’s
tidy.
There are three interrelated rules that make a dataset tidy:
- Each variable is a column; each column is a variable.
- Each observation is a row; each row is an observation.
- Each value is a cell; each cell is a single value.

Why ensure that your data is tidy? There are two main advantages:
There’s a general advantage to picking one consistent way of storing data. If you have a consistent data structure, it’s easier to learn the tools that work with it because they have an underlying uniformity.
There’s a specific advantage to placing variables in columns because it allows Polar’s vectorized nature to shine. Plotly also works by default with tidy data formats, and requires (a little) workaround for wide formats.
Here are some examples of how Polars & Plotly work with tidy data:
# Compute rate per 10,000
table1.with_columns(
rate = pl.col("cases") / pl.col("population") * 1000
)| country | year | cases | population | rate |
|---|---|---|---|---|
| str | i64 | i64 | i64 | f64 |
| "Afghanistan" | 1999 | 745 | 19987071 | 0.037274 |
| "Afghanistan" | 2000 | 2666 | 20595360 | 0.129447 |
| "Brazil" | 1999 | 37737 | 172006362 | 0.219393 |
| "Brazil" | 2000 | 80488 | 174504898 | 0.461236 |
| "China" | 1999 | 212258 | 1272915272 | 0.16675 |
| "China" | 2000 | 213766 | 1280428583 | 0.166949 |
# Compute total cases per year
table1.group_by("year").agg(total_cases = pl.col("cases").sum())| year | total_cases |
|---|---|
| i64 | i64 |
| 1999 | 250740 |
| 2000 | 296920 |
# Visualize changes over time
table1_datefix = table1.with_columns(
date = (pl.col("year").cast(pl.String) + "-01-01").str.to_date()
)
px.line(
table1_datefix,
x="date",
y="cases",
color="country",
symbol="country",
title="Cases by Year and Country",
)While we are covering the basics of tidy data, I highly recommend reading Tidy Data by Hadley Wickham. It’s a short paper on the principles of working with tidy data and it’s benefits.
4.2 Lengthening data
The principles of tidy data might seem so obvious that you wonder if you’ll ever encounter a dataset that isn’t tidy. Unfortunately, however, most real data is untidy. There are two main reasons:
Data is often organized to facilitate some goal other than analysis. For example, it’s common for data to be structured to make data entry, not analysis, easy.
Most people aren’t familiar with the principles of tidy data, and it’s hard to derive them yourself unless you spend a lot of time working with data.
This means that most real analyses will require at least a little tidying. You’ll begin by figuring out what the underlying variables and observations are. Sometimes this is easy; other times you’ll need to consult with the people who originally generated the data. Next, you’ll pivot your data into a tidy form, with variables in the columns and observations in the rows.
Polars provides two methods for pivoting data:
DataFrame.pivot() and DataFrame.unpivot().
We’ll first start with DataFrame.unpivot() because it’s the
most common case. Let’s dive into some examples.
4.2.1 Data in column names
The billboard dataset records the billboard rank of
songs in the year 2000:
billboard = pl.read_csv("data/billboard.csv", try_parse_dates=True)
billboard| artist | track | date.entered | wk1 | wk2 | wk3 | wk4 | wk5 | wk6 | wk7 | wk8 | wk9 | wk10 | wk11 | wk12 | wk13 | wk14 | wk15 | wk16 | wk17 | wk18 | wk19 | wk20 | wk21 | wk22 | wk23 | wk24 | wk25 | wk26 | wk27 | wk28 | wk29 | wk30 | wk31 | wk32 | wk33 | wk34 | … | wk40 | wk41 | wk42 | wk43 | wk44 | wk45 | wk46 | wk47 | wk48 | wk49 | wk50 | wk51 | wk52 | wk53 | wk54 | wk55 | wk56 | wk57 | wk58 | wk59 | wk60 | wk61 | wk62 | wk63 | wk64 | wk65 | wk66 | wk67 | wk68 | wk69 | wk70 | wk71 | wk72 | wk73 | wk74 | wk75 | wk76 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| str | str | date | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | … | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | str | str | str | str | str | str | str | str | str | str | str |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | 87 | 82 | 72 | 77 | 87 | 94 | 99 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | … | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "2Ge+her" | "The Hardest Part Of ..." | 2000-09-02 | 91 | 87 | 92 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | … | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "3 Doors Down" | "Kryptonite" | 2000-04-08 | 81 | 70 | 68 | 67 | 66 | 57 | 54 | 53 | 51 | 51 | 51 | 51 | 47 | 44 | 38 | 28 | 22 | 18 | 18 | 14 | 12 | 7 | 6 | 6 | 6 | 5 | 5 | 4 | 4 | 4 | 4 | 3 | 3 | 3 | … | 15 | 14 | 13 | 14 | 16 | 17 | 21 | 22 | 24 | 28 | 33 | 42 | 42 | 49 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "3 Doors Down" | "Loser" | 2000-10-21 | 76 | 76 | 72 | 69 | 67 | 65 | 55 | 59 | 62 | 61 | 61 | 59 | 61 | 66 | 72 | 76 | 75 | 67 | 73 | 70 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | … | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| "Yearwood, Trisha" | "Real Live Woman" | 2000-04-01 | 85 | 83 | 83 | 82 | 81 | 91 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | … | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Ying Yang Twins" | "Whistle While You Tw..." | 2000-03-18 | 95 | 94 | 91 | 85 | 84 | 78 | 74 | 78 | 85 | 89 | 97 | 96 | 99 | 99 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | … | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Zombie Nation" | "Kernkraft 400" | 2000-09-02 | 99 | 99 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | … | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "matchbox twenty" | "Bent" | 2000-04-29 | 60 | 37 | 29 | 24 | 22 | 21 | 18 | 16 | 13 | 12 | 8 | 6 | 1 | 2 | 3 | 2 | 2 | 3 | 4 | 5 | 4 | 4 | 6 | 9 | 12 | 13 | 19 | 20 | 20 | 24 | 29 | 28 | 27 | 30 | … | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
In this dataset, each observation is a song. The first three columns
(artist, track, and date.entered)
are variables that describe the song. Then we have 76 columns,
wk1-wk76, that describe the rank of the song
in each week. Here, the column names are one variable (the
week) and the cell values are another (the
rank).
To tidy this data, we’ll use DataFrame.unpivot():
billboard.unpivot(
index=["artist", "track", "date.entered"],
variable_name="week",
value_name="rank"
)| artist | track | date.entered | week | rank |
|---|---|---|---|---|
| str | str | date | str | str |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk1" | "87" |
| "2Ge+her" | "The Hardest Part Of ..." | 2000-09-02 | "wk1" | "91" |
| "3 Doors Down" | "Kryptonite" | 2000-04-08 | "wk1" | "81" |
| "3 Doors Down" | "Loser" | 2000-10-21 | "wk1" | "76" |
| … | … | … | … | … |
| "Yearwood, Trisha" | "Real Live Woman" | 2000-04-01 | "wk76" | null |
| "Ying Yang Twins" | "Whistle While You Tw..." | 2000-03-18 | "wk76" | null |
| "Zombie Nation" | "Kernkraft 400" | 2000-09-02 | "wk76" | null |
| "matchbox twenty" | "Bent" | 2000-04-29 | "wk76" | null |
There are three key arguments:
indexspecifies which columns are not pivoted, i.e. which columns are variables.variable_namenames the variable stored in the column names, we named that variableweek.value_namenames the variable stored in the cell values, we named that variablerank.
Now let’s turn our attention to the resulting, longer data frame. What happens if a song is in the top 100 for less than 76 weeks? Take 2 Pack’s “Baby Don’t Cry”, for example:
billboard.unpivot(
index=["artist", "track", "date.entered"],
variable_name="week",
value_name="rank"
).filter(
pl.col("artist") == "2 Pac", # using commas is another way to chain multiple ANDs
pl.col("track").str.starts_with("Baby Don't Cry")
)| artist | track | date.entered | week | rank |
|---|---|---|---|---|
| str | str | date | str | str |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk1" | "87" |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk2" | "82" |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk3" | "72" |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk4" | "77" |
| … | … | … | … | … |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk73" | null |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk74" | null |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk75" | null |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk76" | null |
The null in the bottom of the table suggests that this
song wasn’t ranked in (at least) weeks 74-76. These nulls
don’t really represent unknown observations; they were forced to exist
by the structure of the dataset, so we can safely filter them out:
billboard.unpivot(
index=["artist", "track", "date.entered"],
variable_name="week",
value_name="rank"
).drop_nulls(
# no arguments will drop any row with `null`
)| artist | track | date.entered | week | rank |
|---|---|---|---|---|
| str | str | date | str | str |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | "wk1" | "87" |
| "2Ge+her" | "The Hardest Part Of ..." | 2000-09-02 | "wk1" | "91" |
| "3 Doors Down" | "Kryptonite" | 2000-04-08 | "wk1" | "81" |
| "3 Doors Down" | "Loser" | 2000-10-21 | "wk1" | "76" |
| … | … | … | … | … |
| "Lonestar" | "Amazed" | 1999-06-05 | "wk63" | "45" |
| "Creed" | "Higher" | 1999-09-11 | "wk64" | "50" |
| "Lonestar" | "Amazed" | 1999-06-05 | "wk64" | "50" |
| "Creed" | "Higher" | 1999-09-11 | "wk65" | "49" |
The number of rows is now much lower, indicating that many rows with
nulls were dropped.
You might also wonder what happens if a song is in the Top 100 for
more than 76 weeks? We can’t tell from this data, but you might guess
that additional columns wk77, wk78,
... would be added to the dataset.
This data is now tidy, but we could make future computation a bit
easier by converting values of week and rank
from character strings to numbers using
DataFrame.with_columns().
billboard_longer = (
billboard
.unpivot(
index=["artist", "track", "date.entered"],
variable_name="week",
value_name="rank"
).drop_nulls(
).with_columns(
pl.col("week").str.extract(r"(\d+)").str.to_integer(),
pl.col("rank").str.to_integer()
)
)
billboard_longer| artist | track | date.entered | week | rank |
|---|---|---|---|---|
| str | str | date | i64 | i64 |
| "2 Pac" | "Baby Don't Cry (Keep..." | 2000-02-26 | 1 | 87 |
| "2Ge+her" | "The Hardest Part Of ..." | 2000-09-02 | 1 | 91 |
| "3 Doors Down" | "Kryptonite" | 2000-04-08 | 1 | 81 |
| "3 Doors Down" | "Loser" | 2000-10-21 | 1 | 76 |
| … | … | … | … | … |
| "Lonestar" | "Amazed" | 1999-06-05 | 63 | 45 |
| "Creed" | "Higher" | 1999-09-11 | 64 | 50 |
| "Lonestar" | "Amazed" | 1999-06-05 | 64 | 50 |
| "Creed" | "Higher" | 1999-09-11 | 65 | 49 |
Now that we have all the week numbers in one variable and all the rank values in another, we’re in a good position to visualize how song ranks vary over time.
px.line(
billboard_longer, x="week", y="rank", line_group="track"
).update_traces( # we'll learn more about these extra methods in a later module
opacity=0.25
).update_yaxes(
autorange="reversed"
)We can see that very few songs stay in the top 100 for more than 20 weeks.
4.2.2 How does pivoting work?
Now that you’ve seen how we can use pivoting to reshape our data,
let’s take a little time to gain some intuition about what pivoting does
to the data. Let’s start with a very simple dataset to make it easier to
see what’s happening. Suppose we have three patients with
ids A, B, and C, and we take two blood pressure
measurements on each patient.
df = pl.from_dict({
"id": ["A", "B", "C"],
"bp1": [100, 140, 120],
"bp2": [120, 114, 125]
})
df| id | bp1 | bp2 |
|---|---|---|
| str | i64 | i64 |
| "A" | 100 | 120 |
| "B" | 140 | 114 |
| "C" | 120 | 125 |
We want our new dataset to have three variables: id
(already exists), measurement (the column names), and
value (the cell values). To achieve this, we need to pivot
df longer:
df.unpivot(
index = "id",
variable_name="measurement",
value_name="value"
).sort(
["id", "measurement", "value"]
)| id | measurement | value |
|---|---|---|
| str | str | i64 |
| "A" | "bp1" | 100 |
| "A" | "bp2" | 120 |
| "B" | "bp1" | 140 |
| "B" | "bp2" | 114 |
| "C" | "bp1" | 120 |
| "C" | "bp2" | 125 |
How does the reshaping work? It’s easier to see if we think about it
column by column. As shown below, the values in a column that was
already a variable in the original dataset (id) need to be
repeated, once for each column that is pivoted.
The column names become values in a new variable, whose name is
defined by variable_name, as shown below. THey need to be
repeated once for each row in the original dataset.
The cell values also become values in a new variable, with a name
defined by values_name. They are unwound row by row, as
shown below.
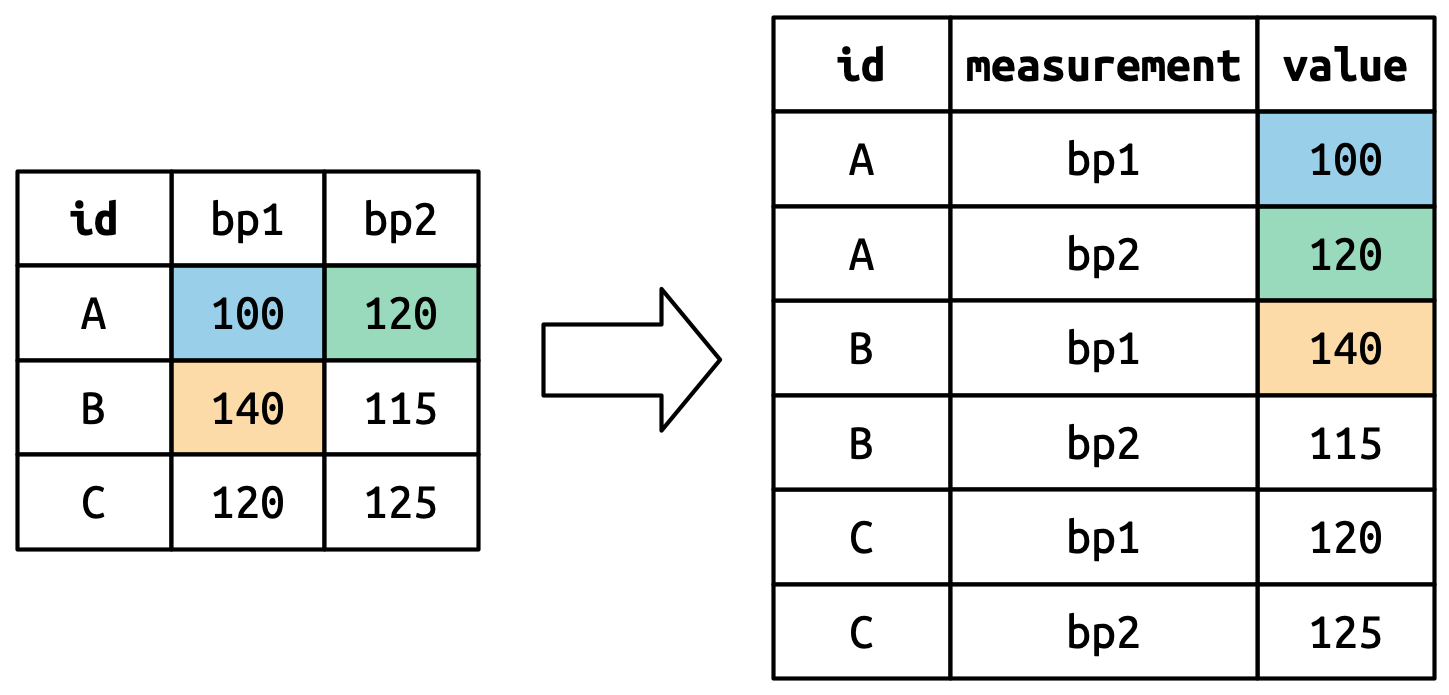
4.2.3 More variables in column names
A more challenging situation occurs when you have multiple pieces of
information crammed into the column names, and you would like to store
these in separate new variables. For example, take the who
dataset, the source of table1 and friends you saw
above:
who2 = pl.read_csv("data/who2.csv", null_values="")
who2| country | year | sp_m_014 | sp_m_1524 | sp_m_2534 | sp_m_3544 | sp_m_4554 | sp_m_5564 | sp_m_65 | sp_f_014 | sp_f_1524 | sp_f_2534 | sp_f_3544 | sp_f_4554 | sp_f_5564 | sp_f_65 | sn_m_014 | sn_m_1524 | sn_m_2534 | sn_m_3544 | sn_m_4554 | sn_m_5564 | sn_m_65 | sn_f_014 | sn_f_1524 | sn_f_2534 | sn_f_3544 | sn_f_4554 | sn_f_5564 | sn_f_65 | ep_m_014 | ep_m_1524 | ep_m_2534 | ep_m_3544 | ep_m_4554 | ep_m_5564 | ep_m_65 | ep_f_014 | ep_f_1524 | ep_f_2534 | ep_f_3544 | ep_f_4554 | ep_f_5564 | ep_f_65 | rel_m_014 | rel_m_1524 | rel_m_2534 | rel_m_3544 | rel_m_4554 | rel_m_5564 | rel_m_65 | rel_f_014 | rel_f_1524 | rel_f_2534 | rel_f_3544 | rel_f_4554 | rel_f_5564 | rel_f_65 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| str | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 | i64 |
| "Afghanistan" | 1980 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Afghanistan" | 1981 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Afghanistan" | 1982 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Afghanistan" | 1983 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … | … |
| "Zimbabwe" | 2010 | 150 | 710 | 2208 | 1682 | 761 | 350 | 252 | 173 | 974 | 2185 | 1283 | 490 | 265 | 171 | 1826 | 821 | 3342 | 3270 | 1545 | 882 | 864 | 1732 | 1282 | 4013 | 2851 | 1377 | 789 | 563 | 270 | 243 | 902 | 868 | 418 | 229 | 192 | 220 | 319 | 1058 | 677 | 338 | 181 | 146 | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Zimbabwe" | 2011 | 152 | 784 | 2467 | 2071 | 780 | 377 | 278 | 174 | 1084 | 2161 | 1386 | 448 | 274 | 160 | 1364 | 596 | 2473 | 2813 | 1264 | 702 | 728 | 1271 | 947 | 2754 | 2216 | 962 | 587 | 495 | 250 | 195 | 746 | 796 | 342 | 172 | 172 | 209 | 318 | 802 | 640 | 284 | 137 | 129 | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Zimbabwe" | 2012 | 120 | 783 | 2421 | 2086 | 796 | 360 | 271 | 173 | 939 | 2053 | 1286 | 483 | 231 | 161 | 1169 | 613 | 2302 | 2657 | 1154 | 708 | 796 | 1008 | 888 | 2287 | 1957 | 829 | 516 | 432 | 233 | 214 | 658 | 789 | 331 | 178 | 182 | 208 | 319 | 710 | 579 | 228 | 140 | 143 | null | null | null | null | null | null | null | null | null | null | null | null | null | null |
| "Zimbabwe" | 2013 | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | null | 1315 | 1642 | 5331 | 5363 | 2349 | 1206 | 1208 | 1252 | 2069 | 4649 | 3526 | 1453 | 811 | 725 |
who2.glimpse()Rows: 7240
Columns: 58
$ country <str> 'Afghanistan', 'Afghanistan', 'Afghanistan', 'Afghanistan', 'Afghanistan', 'Afghanistan', 'Afghanistan', 'Afghanistan', 'Afghanistan', 'Afghanistan'
$ year <i64> 1980, 1981, 1982, 1983, 1984, 1985, 1986, 1987, 1988, 1989
$ sp_m_014 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_m_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_m_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_m_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_m_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_m_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_m_65 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_f_014 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_f_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_f_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_f_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_f_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_f_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ sp_f_65 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_m_014 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_m_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_m_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_m_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_m_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_m_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_m_65 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_f_014 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_f_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_f_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_f_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_f_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_f_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ sn_f_65 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_m_014 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_m_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_m_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_m_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_m_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_m_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_m_65 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_f_014 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_f_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_f_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_f_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_f_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_f_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ ep_f_65 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_m_014 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_m_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_m_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_m_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_m_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_m_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_m_65 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_f_014 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_f_1524 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_f_2534 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_f_3544 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_f_4554 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_f_5564 <i64> None, None, None, None, None, None, None, None, None, None
$ rel_f_65 <i64> None, None, None, None, None, None, None, None, None, None
This dataset, collected by the World Health Organization, records
information about tuberculosis diagnoses. There are two columns that are
already variables and are easy to interpret: country and
year. They are followed by 56 columns like
sp_m_014, ep_m_4554, and
rel_m_3544. If you stare at these columns for long enough,
you’ll notice there’s a pattern. Each column name is made up of three
pieces separated by _. The first piece,
sp/rel/ep, describes the method
used for the diagnosis, the second piece, m/f
is the gender (coded as a binary variable in this dataset), and the
third piece,
014/1524/2534/3544/4554/5564/65
is the age range (014 represents 0-14, for example).
So in this case, we have six pieces of information recorded in
who2: the country and the year (already columns); the
method of diagnosis, the gender category, and the age range category
(contained in the other column names); and the count of patients in that
category (cell values).
To organize these six pieces of information in six separate columns,
first we use DataFrame.unpivot() like before, then we have
to do some data wrangling using list
expressions to split the information from the column names into
separate columns:
- Unpivot:
step1 = who2.unpivot(
index=["country", "year"],
variable_name="key",
value_name="count"
)
step1| country | year | key | count |
|---|---|---|---|
| str | i64 | str | i64 |
| "Afghanistan" | 1980 | "sp_m_014" | null |
| "Afghanistan" | 1981 | "sp_m_014" | null |
| "Afghanistan" | 1982 | "sp_m_014" | null |
| "Afghanistan" | 1983 | "sp_m_014" | null |
| … | … | … | … |
| "Zimbabwe" | 2010 | "rel_f_65" | null |
| "Zimbabwe" | 2011 | "rel_f_65" | null |
| "Zimbabwe" | 2012 | "rel_f_65" | null |
| "Zimbabwe" | 2013 | "rel_f_65" | 725 |
- Split the
keycolumn intodiagnosis,gender, andagecolumns:
step2 = step1.with_columns(
pl.col("key").str.split("_")
)
step2| country | year | key | count |
|---|---|---|---|
| str | i64 | list[str] | i64 |
| "Afghanistan" | 1980 | ["sp", "m", "014"] | null |
| "Afghanistan" | 1981 | ["sp", "m", "014"] | null |
| "Afghanistan" | 1982 | ["sp", "m", "014"] | null |
| "Afghanistan" | 1983 | ["sp", "m", "014"] | null |
| … | … | … | … |
| "Zimbabwe" | 2010 | ["rel", "f", "65"] | null |
| "Zimbabwe" | 2011 | ["rel", "f", "65"] | null |
| "Zimbabwe" | 2012 | ["rel", "f", "65"] | null |
| "Zimbabwe" | 2013 | ["rel", "f", "65"] | 725 |
- Extract each list element into a new column:
step3 = step2.select(
pl.col("country"),
pl.col("year"),
pl.col("key").list.get(0).alias("diagnosis"), # Polars also uses 0-indexing
pl.col("key").list.get(1).alias("gender"),
pl.col("key").list.get(2).alias("age"),
pl.col("count")
)
step3| country | year | diagnosis | gender | age | count |
|---|---|---|---|---|---|
| str | i64 | str | str | str | i64 |
| "Afghanistan" | 1980 | "sp" | "m" | "014" | null |
| "Afghanistan" | 1981 | "sp" | "m" | "014" | null |
| "Afghanistan" | 1982 | "sp" | "m" | "014" | null |
| "Afghanistan" | 1983 | "sp" | "m" | "014" | null |
| … | … | … | … | … | … |
| "Zimbabwe" | 2010 | "rel" | "f" | "65" | null |
| "Zimbabwe" | 2011 | "rel" | "f" | "65" | null |
| "Zimbabwe" | 2012 | "rel" | "f" | "65" | null |
| "Zimbabwe" | 2013 | "rel" | "f" | "65" | 725 |
All together:
(
who2.unpivot(
index=["country", "year"], variable_name="key", value_name="count"
)
.with_columns(
pl.col("key").str.split("_")
)
.select(
pl.col("country"),
pl.col("year"),
pl.col("key").list.get(0).alias("diagnosis"),
pl.col("key").list.get(1).alias("gender"),
pl.col("key").list.get(2).alias("age"),
pl.col("count")
)
)While the above steps break down the process to understand easier, here is a more concise, albeit more advanced, way to achieve the same results:
(
who2.unpivot(
index=["country", "year"],
variable_name="key",
value_name="count"
)
.with_columns(
pl.col("key")
.str.split("_")
.list.to_struct(fields=["diagnosis", "gender", "age"])
)
.unnest("key")
)| country | year | diagnosis | gender | age | count |
|---|---|---|---|---|---|
| str | i64 | str | str | str | i64 |
| "Afghanistan" | 1980 | "sp" | "m" | "014" | null |
| "Afghanistan" | 1981 | "sp" | "m" | "014" | null |
| "Afghanistan" | 1982 | "sp" | "m" | "014" | null |
| "Afghanistan" | 1983 | "sp" | "m" | "014" | null |
| … | … | … | … | … | … |
| "Zimbabwe" | 2010 | "rel" | "f" | "65" | null |
| "Zimbabwe" | 2011 | "rel" | "f" | "65" | null |
| "Zimbabwe" | 2012 | "rel" | "f" | "65" | null |
| "Zimbabwe" | 2013 | "rel" | "f" | "65" | 725 |
Conceptually, this is only a minor variation on the simpler case you’ve already seen. The figure below shows the basic idea: now, instead of the column names pivoting into a single column, they pivot into multiple columns.
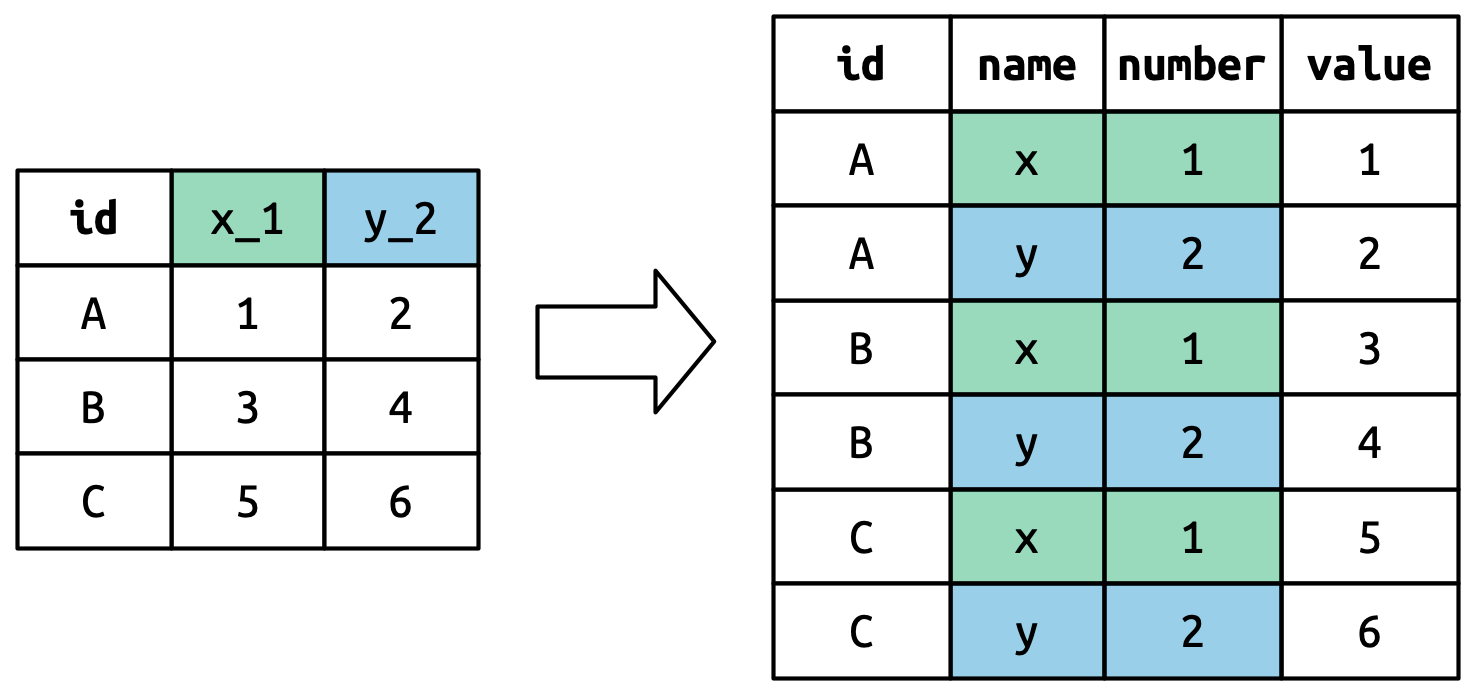
4.3 Widening data
So far we’ve used DataFrame.unpivot() to solve the
common class of problems where values have ended up in column names.
Next we’ll pivot (HA HA) to DataFrame.pivot(), which makes
datasets wider by increasing columns and reducing rows
and helps when one observation is spread across multiple rows. This
seems to arise less commonly in the wild, but it does seem to crop up a
lot when dealing with governmental data.
We’ll start by looking at cms_patient_experience, a
dataset from the Centers of Medicare and Medicaid services that collects
data about patient experiences:
cms_patient_experience = pl.read_csv(
"data/cms_patient_experience.csv",
schema_overrides={"org_pac_id": pl.String}
)
cms_patient_experience| org_pac_id | org_nm | measure_cd | measure_title | prf_rate |
|---|---|---|---|---|
| str | str | str | str | i64 |
| "0446157747" | "USC CARE MEDICAL GROUP INC" | "CAHPS_GRP_1" | "CAHPS for MIPS SSM: Getting Ti… | 63 |
| "0446157747" | "USC CARE MEDICAL GROUP INC" | "CAHPS_GRP_2" | "CAHPS for MIPS SSM: How Well P… | 87 |
| "0446157747" | "USC CARE MEDICAL GROUP INC" | "CAHPS_GRP_3" | "CAHPS for MIPS SSM: Patient's … | 86 |
| "0446157747" | "USC CARE MEDICAL GROUP INC" | "CAHPS_GRP_5" | "CAHPS for MIPS SSM: Health Pro… | 57 |
| … | … | … | … | … |
| "9931011434" | "PATIENT FIRST RICHMOND MEDICAL… | "CAHPS_GRP_2" | "CAHPS for MIPS SSM: How Well P… | null |
| "9931011434" | "PATIENT FIRST RICHMOND MEDICAL… | "CAHPS_GRP_3" | "CAHPS for MIPS SSM: Patient's … | null |
| "9931011434" | "PATIENT FIRST RICHMOND MEDICAL… | "CAHPS_GRP_5" | "CAHPS for MIPS SSM: Health Pro… | 45 |
| "9931011434" | "PATIENT FIRST RICHMOND MEDICAL… | "CAHPS_GRP_12" | "CAHPS for MIPS SSM: Stewardshi… | 19 |
The core unit being studied is an organization, but each organization
is spread across six rows, with one row for each measurement taken in
the survey organization. We can see the complete set of values for
measure_cd and measure_title by using
DataFrame.unique().select():
cols = ["measure_cd", "measure_title"]
cms_patient_experience.unique(subset=cols).select(cols).sort(cols)| measure_cd | measure_title |
|---|---|
| str | str |
| "CAHPS_GRP_1" | "CAHPS for MIPS SSM: Getting Ti… |
| "CAHPS_GRP_12" | "CAHPS for MIPS SSM: Stewardshi… |
| "CAHPS_GRP_2" | "CAHPS for MIPS SSM: How Well P… |
| "CAHPS_GRP_3" | "CAHPS for MIPS SSM: Patient's … |
| "CAHPS_GRP_5" | "CAHPS for MIPS SSM: Health Pro… |
| "CAHPS_GRP_8" | "CAHPS for MIPS SSM: Courteous … |
Neither of these columns will make particularly great variable names:
measure_cd doesn’t hint at the meaning of the variable and
measure_title is a long sentence containing spaces. We’ll
use measure_cd as the source for our new column names for
now, but in a real analysis you might want to create your own variable
names that are both short and meaningful.
DataFrame.pivot() has a similar, but different,
interface than DataFrame.unpivot(): you still select the
index columns (the columns that remain), but now you select
which column the values will come from, and which column
the names come from (on).
cms_patient_experience.pivot(
index=["org_pac_id", "org_nm"],
on=["measure_cd"], # Notice we don't use `measure_title`
values="prf_rate"
)| org_pac_id | org_nm | CAHPS_GRP_1 | CAHPS_GRP_2 | CAHPS_GRP_3 | CAHPS_GRP_5 | CAHPS_GRP_8 | CAHPS_GRP_12 |
|---|---|---|---|---|---|---|---|
| str | str | i64 | i64 | i64 | i64 | i64 | i64 |
| "0446157747" | "USC CARE MEDICAL GROUP INC" | 63 | 87 | 86 | 57 | 85 | 24 |
| "0446162697" | "ASSOCIATION OF UNIVERSITY PHYS… | 59 | 85 | 83 | 63 | 88 | 22 |
| "0547164295" | "BEAVER MEDICAL GROUP PC" | 49 | null | 75 | 44 | 73 | 12 |
| "0749333730" | "CAPE PHYSICIANS ASSOCIATES PA" | 67 | 84 | 85 | 65 | 82 | 24 |
| … | … | … | … | … | … | … | … |
| "9830008770" | "WASHINGTON UNIVERSITY" | 65 | null | null | 61 | null | 28 |
| "9830093640" | "COOPERATIVE HEALTHCARE SERVICE… | 65 | 87 | 78 | 52 | null | 25 |
| "9830094515" | "SUTTER VALLEY MEDICAL FOUNDATI… | 60 | null | 88 | 55 | null | 24 |
| "9931011434" | "PATIENT FIRST RICHMOND MEDICAL… | 80 | null | null | 45 | null | 19 |
4.3.1
How does DataFrame.pivot() work?
To understand how DataFrame.pivot() works, let’s again
start with a very simple dataset. This time we have two patients with
ids A and B, we have three blood pressure measurements on
patient A and two on patient B:
df = pl.from_dict({
"id": ["A", "B", "B", "A", "A"],
"measurement": ["bp1", "bp1", "bp2", "bp2", "bp3"],
"value": [100, 140, 115, 120, 105]
})
df| id | measurement | value |
|---|---|---|
| str | str | i64 |
| "A" | "bp1" | 100 |
| "B" | "bp1" | 140 |
| "B" | "bp2" | 115 |
| "A" | "bp2" | 120 |
| "A" | "bp3" | 105 |
We’ll take the values from the value column and the
names from the measurement column:
df.pivot(on="measurement", values="value") # unused cols get put in `index`| id | bp1 | bp2 | bp3 |
|---|---|---|---|
| str | i64 | i64 | i64 |
| "A" | 100 | 120 | 105 |
| "B" | 140 | 115 | null |
To begin the process, DataFrame.pivot() needs to first
figure out what will go in the rows and columns. The new column names
will be the unique values of measurement.
df.select("measurement").unique()| measurement |
|---|
| str |
| "bp2" |
| "bp3" |
| "bp1" |
By default, the rows in the output are determined by all the
variables that aren’t going into the new names or values. These are
called the index columns. Here there is only one column,
but in general there can be any number.
df.select(pl.exclude("measurement", "value")).unique()| id |
|---|
| str |
| "A" |
| "B" |
DataFrame.pivot() then combines these results to
generate an empty data frame:
df.select(pl.exclude("measurement", "value")).unique().with_columns(
x=pl.lit(None), y=pl.lit(None), z=pl.lit(None)
)| id | x | y | z |
|---|---|---|---|
| str | null | null | null |
| "A" | null | null | null |
| "B" | null | null | null |
It then fills in all the missing values using the data in the input. In this case, not every cell in the output has a corresponding value in the input as there’s no third blood pressure measurement for patient B, so that cell remains missing.
4.3.2 Data and variable names in the column headers
We will now see an example where we need to unpivot and pivot in the
same set of steps to tidy our data. This step up in complexity is when
the column names include a mix of variable values and variable names.
For example, take the household dataset:
household = pl.from_dict({
"family": [1, 2, 3, 4, 5],
"dob_child1": ["1998-11-26", "1996-06-22", "2002-07-11", "2004-10-10", "2000-12-05"],
"dob_child2": ["2000-01-29", None, "2004-04-05", "2009-08-27", "2005-02-28"],
"name_child1": ["Susan", "Mark", "Sam", "Craig", "Parker"],
"name_child2": ["Jose", None, "Seth", "Khai", "Gracie"]
}).with_columns(
cs.starts_with("dob").str.to_date()
)
household| family | dob_child1 | dob_child2 | name_child1 | name_child2 |
|---|---|---|---|---|
| i64 | date | date | str | str |
| 1 | 1998-11-26 | 2000-01-29 | "Susan" | "Jose" |
| 2 | 1996-06-22 | null | "Mark" | null |
| 3 | 2002-07-11 | 2004-04-05 | "Sam" | "Seth" |
| 4 | 2004-10-10 | 2009-08-27 | "Craig" | "Khai" |
| 5 | 2000-12-05 | 2005-02-28 | "Parker" | "Gracie" |
This dataset contains data about five families, with the names and
dates of birth of up to two children. The new challenge in this dataset
is that the column names contain the names of two variables
(dob, name) and the values of another
(child, with values 1 or 2).
(
# First, unpivot the data frame to create rows for each family-column combination
household.unpivot(index="family")
# Extract the base column name (dob/name) and the child number
.with_columns(
pl.col("variable")
.str.split("_")
.list.to_struct(fields=["base_col", "child"])
)
.unnest("variable")
# Pivot the data to create separate columns for each base_col (dob, name)
.pivot(index=["family", "child"], on="base_col", values="value")
# Filter out rows with null values
.drop_nulls()
# Clean up results
.sort(["family", "child"])
.with_columns(pl.col("dob").str.to_date())
)| family | child | dob | name |
|---|---|---|---|
| i64 | str | date | str |
| 1 | "child1" | 1998-11-26 | "Susan" |
| 1 | "child2" | 2000-01-29 | "Jose" |
| 2 | "child1" | 1996-06-22 | "Mark" |
| 3 | "child1" | 2002-07-11 | "Sam" |
| … | … | … | … |
| 4 | "child1" | 2004-10-10 | "Craig" |
| 4 | "child2" | 2009-08-27 | "Khai" |
| 5 | "child1" | 2000-12-05 | "Parker" |
| 5 | "child2" | 2005-02-28 | "Gracie" |
We use DataFrame.drop_nulls() since the shape of the
input forces the creation of explicit missing variables (e.g., for
families that only have one child).
4.4 Summary
In this section, you learned about tidy data: data that has variables
in columns and observations in rows. Tidy data makes working in the
Polars easier, because it’s a consistent structure understood by most
functions, the main challenge is transforming the data from whatever
structure you receive it in to a tidy format. To that end, you learned
about DataFrame.unpivot() and
DataFrame.pivot() which allow you to tidy up many untidy
datasets.
Another challenge is that, for a given dataset, it can be impossible to label the longer or the wider version as the “tidy” one. This is partly a reflection of our definition of tidy data, where we said tidy data has one variable in each column, but we didn’t actually define what a variable is (and it’s surprisingly hard to do so). It’s totally fine to be pragmatic and to say a variable is whatever makes your analysis easiest. So if you’re stuck figuring out how to do some computation, consider switching up the organization of your data; don’t be afraid to untidy, transform, and re-tidy as needed!
5 Data import & export
Working with data provided by Python packages is a great way to learn data science tools, but at some point, you’ll want to apply what you’ve learned to your own data. In this section, you’ll learn the basics of reading data files into Python and how to export them for others (or yourself in the future) to use.
Specifically, this chapter will focus on reading plain-text rectangular files. We’ll start with practical advice for handling features like column names, types, and missing data. You will then learn about reading data from multiple files at once and writing data from Python to a file. Finally, you’ll learn how to handcraft data frames in Python, and export them with Polars functions.
import io # we'll use this to "create" CSVs within Python examples5.1 Reading data from a file
To begin, we’ll focus on the most common rectangular data file type: CSV, which is short for comma-separated values. Here is what a simple CSV file looks like. The first row, commonly called the header row, gives the column names, and the following six rows provide the data. The columns are separated, aka delimited, by commas.
Student ID,Full Name,favourite.food,mealPlan,AGE
1,Sunil Huffmann,Strawberry yoghurt,Lunch only,4
2,Barclay Lynn,French fries,Lunch only,5
3,Jayendra Lyne,N/A,Breakfast and lunch,7
4,Leon Rossini,Anchovies,Lunch only,
5,Chidiegwu Dunkel,Pizza,Breakfast and lunch,five
6,Güvenç Attila,Ice cream,Lunch only,6The following table shows a representation of the same data as a table:
| Student ID | Full Name | favourite.food | mealPlan | AGE |
|---|---|---|---|---|
| 1 | Sunil Huffmann | Strawberry yoghurt | Lunch only | 4 |
| 2 | Barclay Lynn | French fries | Lunch only | 5 |
| 3 | Jayendra Lyne | N/A | Breakfast and lunch | 7 |
| 4 | Leon Rossini | Anchovies | Lunch only | NULL |
| 5 | Chidiegwu Dunkel | Pizza | Breakfast and lunch | five |
| 6 | Guvenc Attila | Ice cream | Lunch only | 6 |
We can read this file into Python using pl.read_csv().
The first argument is the most important: the path to the file. You can
think about the path as the address of the file: the file is called
students.csv and it lives in the data
folder.
students = pl.read_csv("data/students.csv")The code above will work if you have the students.csv file in a data folder in your project. You can also read it directly from a URL:
students = pl.read_csv("https://raw.githubusercontent.com/hadley/r4ds/main/data/students.csv")When you run pl.read_csv(), Polars will scan the file
and automatically infer column types based on the data it contains.
Polars doesn’t print a detailed message about column specifications by
default, but you can examine the schema to see the inferred types:
students.schema.to_frame()| Student ID | Full Name | favourite.food | mealPlan | AGE |
|---|---|---|---|---|
| i64 | str | str | str | str |
5.1.1 Practical advice
Once you read data in, the first step usually involves transforming it in some way to make it easier to work with in the rest of your analysis. Let’s take another look at the students data with that in mind.
students| Student ID | Full Name | favourite.food | mealPlan | AGE |
|---|---|---|---|---|
| i64 | str | str | str | str |
| 1 | "Sunil Huffmann" | "Strawberry yoghurt" | "Lunch only" | "4" |
| 2 | "Barclay Lynn" | "French fries" | "Lunch only" | "5" |
| 3 | "Jayendra Lyne" | "N/A" | "Breakfast and lunch" | "7" |
| 4 | "Leon Rossini" | "Anchovies" | "Lunch only" | null |
| 5 | "Chidiegwu Dunkel" | "Pizza" | "Breakfast and lunch" | "five" |
| 6 | "Güvenç Attila" | "Ice cream" | "Lunch only" | "6" |
In the favourite.food column, there’s an entry “N/A”
that should be interpreted as a real None that Python will
recognize as “not available”. This is something we can address using the
null_values argument. By default, Polars only recognizes
empty strings and the string “null” (case-insensitive) as missing
values.
students = pl.read_csv(
"data/students.csv",
null_values=["N/A", ""]
)
students| Student ID | Full Name | favourite.food | mealPlan | AGE |
|---|---|---|---|---|
| i64 | str | str | str | str |
| 1 | "Sunil Huffmann" | "Strawberry yoghurt" | "Lunch only" | "4" |
| 2 | "Barclay Lynn" | "French fries" | "Lunch only" | "5" |
| 3 | "Jayendra Lyne" | null | "Breakfast and lunch" | "7" |
| 4 | "Leon Rossini" | "Anchovies" | "Lunch only" | null |
| 5 | "Chidiegwu Dunkel" | "Pizza" | "Breakfast and lunch" | "five" |
| 6 | "Güvenç Attila" | "Ice cream" | "Lunch only" | "6" |
You might also notice that Student ID and
Full Name columns contain spaces, which can make them
awkward to work with in code. In Polars, you can refer to these columns
using either bracket notation or with the . accessor, but
the latter won’t work with spaces or special characters:
# Works with spaces
first_student_name = students["Full Name"][0]
print(first_student_name)
# Won't work with spaces
first_student_name = students.Full Name[0] # Syntax error!Cell In[94], line 6 first_student_name = students.Full Name[0] # Syntax error! ^ SyntaxError: invalid syntax
It’s often a good idea to rename these columns to follow Python naming conventions. Here’s how you can rename specific columns:
students = students.rename({
"Student ID": "student_id",
"Full Name": "full_name"
})
students| student_id | full_name | favourite.food | mealPlan | AGE |
|---|---|---|---|---|
| i64 | str | str | str | str |
| 1 | "Sunil Huffmann" | "Strawberry yoghurt" | "Lunch only" | "4" |
| 2 | "Barclay Lynn" | "French fries" | "Lunch only" | "5" |
| 3 | "Jayendra Lyne" | null | "Breakfast and lunch" | "7" |
| 4 | "Leon Rossini" | "Anchovies" | "Lunch only" | null |
| 5 | "Chidiegwu Dunkel" | "Pizza" | "Breakfast and lunch" | "five" |
| 6 | "Güvenç Attila" | "Ice cream" | "Lunch only" | "6" |
An alternative approach is to use a function to clean all column names at once. Polars doesn’t have a built-in function for this, but we can use a special shortcut called a lambda function to do this in one line. Think of a lambda function as a mini-function that you can write directly where you need it, without having to define it separately. It’s like giving Polars quick instructions on how to transform each column name.
students = (
students.rename(
lambda col_name: col_name.lower().replace(" ", "_").replace(".", "_")
)
)With this code, we are telling Polars to take each column
name (not the values of the columns), call it col,
and do these three things to it:
- Convert it to lowercase
- Replace any spaces with underscores
- Replace any periods with underscores
Note that these are the string methods we learned earlier in this
course, not functions that come from Polars. col_name has
the type str.
Like in for loops, it doesn’t matter what the looping
(or lambda) variable is called. I just called it col_name
because that is what we are effecting. I could’ve also called
it x and it would work the same:
students = (
students.rename(
lambda x: x.lower().replace(" ", "_").replace(".", "_")
)
)If you wanted to use a regular function, you could do something like this:
def clean_col_name(col_name):
return col_name.lower().replace(" ", "_").replace(".", "_")
students = students.rename(clean_column_name)
# Note: no parentheses here, we're passing the function itselfWhen would you want to use a named/user-defined function over a lambda function? It comes down to how many times you would repeat the steps. If you are working with only one data frame, a lambda function might be quicker to write, but if you are going to clean multiple data frames in the same script, creating a named function to use over again would be the better method.
You can use this code snippet to help you with your labs and tests.
You don’t need to fully understand lambda functions for this course, but
you can think of this specific pattern as a helpful ‘recipe’ for
cleaning column names in one step while keeping your code clean and
readable. If you ever need to customize how column names are cleaned,
you can just adjust what happens inside the lambda function. The Polars
documentation
for DataFrame.rename() has another example using lambda
functions to rename columns.
Another common task after reading in data is to consider variable
types. For example, meal_plan is a categorical variable
with a known set of possible values, which in Python should be
represented as a category:
students = students.with_columns(
pl.col("mealplan").cast(pl.Categorical)
)
students| student_id | full_name | favourite_food | mealplan | age |
|---|---|---|---|---|
| i64 | str | str | cat | str |
| 1 | "Sunil Huffmann" | "Strawberry yoghurt" | "Lunch only" | "4" |
| 2 | "Barclay Lynn" | "French fries" | "Lunch only" | "5" |
| 3 | "Jayendra Lyne" | null | "Breakfast and lunch" | "7" |
| 4 | "Leon Rossini" | "Anchovies" | "Lunch only" | null |
| 5 | "Chidiegwu Dunkel" | "Pizza" | "Breakfast and lunch" | "five" |
| 6 | "Güvenç Attila" | "Ice cream" | "Lunch only" | "6" |
Before you analyze this dataset, you’ll probably want to fix the age
column. Currently, age contains mixed types because one
observation is typed out as “five” instead of the number 5:
students = students.with_columns(
age=pl.when(pl.col("age") == "five")
.then(pl.lit(5))
.otherwise(pl.col("age").cast(pl.Int64, strict=False))
) # This is conceptually very similar to Python's built in if-elif-else
students| student_id | full_name | favourite_food | mealplan | age |
|---|---|---|---|---|
| i64 | str | str | cat | i64 |
| 1 | "Sunil Huffmann" | "Strawberry yoghurt" | "Lunch only" | 4 |
| 2 | "Barclay Lynn" | "French fries" | "Lunch only" | 5 |
| 3 | "Jayendra Lyne" | null | "Breakfast and lunch" | 7 |
| 4 | "Leon Rossini" | "Anchovies" | "Lunch only" | null |
| 5 | "Chidiegwu Dunkel" | "Pizza" | "Breakfast and lunch" | 5 |
| 6 | "Güvenç Attila" | "Ice cream" | "Lunch only" | 6 |
The Polars code above uses a conditional expression with
when(), then(), and otherwise()
to handle the special case, then attempts to cast the column to a
floating-point number. The strict=False parameter tells
Polars to convert values that can be parsed as numbers and set the rest
to None.
5.1.2 Other arguments
Usually, pl.read_csv() uses the first line of the data
for the column names, which is a very common convention. But it’s not
uncommon for a few lines of metadata to be included at the top of the
file. You can use skip_rows to skip the first n lines:
csv_data = """The first line of metadata
The second line of metadata
x,y,z
1,2,3"""
pl.read_csv(io.StringIO(csv_data), skip_rows=2)| x | y | z |
|---|---|---|
| i64 | i64 | i64 |
| 1 | 2 | 3 |
In other cases, the data might not have column names. You can use
has_header=False to tell Polars not to treat the first row
as headings:
csv_data = """1,2,3
4,5,6"""
pl.read_csv(io.StringIO(csv_data), has_header=False)| column_1 | column_2 | column_3 |
|---|---|---|
| i64 | i64 | i64 |
| 1 | 2 | 3 |
| 4 | 5 | 6 |
By default, Polars will name the columns “column_1”, “column_2”, etc.
You can provide your own column names with the new_columns
parameter:
csv_data = """1,2,3
4,5,6"""
pl.read_csv(
io.StringIO(csv_data),
has_header=False,
new_columns=["x", "y", "z"]
)| x | y | z |
|---|---|---|
| i64 | i64 | i64 |
| 1 | 2 | 3 |
| 4 | 5 | 6 |
These arguments are all you need to know to read the majority of CSV
files that you’ll encounter in practice. (For the rest, you’ll need to
carefully inspect your .csv file and read the documentation for
pl.read_csv()’s many other arguments.)
5.1.3 Other file types
Once you’ve mastered read_csv(), using Polars’ other
functions is straightforward; it’s just a matter of knowing which
function to reach for:
read_csv()reads comma-separated files.read_csv()can also read semicolon-separated files by settingseparator=";".read_csv()can read tab-delimited files by settingseparator="\t", or you can use the aliasread_tsv().read_csv()can read files with any delimiter by setting theseparatorparameter.read_ndjson()reads newline-delimited JSON files.read_parquet()reads Apache Parquet files, a columnar storage format that is typically faster and more space-efficient than CSV.read_ipc()orread_arrow()reads Arrow IPC files, which provide high-performance interoperability between different systems.read_excel()reads Excel files (learn more about the different Excel features here).read_avro()reads Avro files.
5.2 Controlling column types
A CSV file doesn’t contain any information about the type of each variable (i.e., whether it’s a boolean, number, string, etc.), so Polars will try to guess the type. This section describes how the guessing process works, how to resolve some common problems that cause it to fail, and how to supply the column types yourself.
5.2.1 Guessing types
Polars uses a heuristic to figure out the column types. By default,
it samples a certain number of rows from the file and tries to infer the
type based on the values it sees. You can control this with the
infer_schema_length parameter.
The type inference generally follows these rules: - If values are “true” or “false” (case-insensitive), it’s a boolean. - If values are all integers, it’s an integer type. - If values have decimals but are all numeric, it’s a floating-point type. - If values match a date or datetime pattern, it’s a date or datetime type. - Otherwise, it’s a string type.
You can see that behavior with a simple example:
csv_data = """logical,numeric,date,string
TRUE,1,2021-01-15,abc
false,4.5,2021-02-15,def
T,Inf,2021-02-16,ghi"""
pl.read_csv(io.StringIO(csv_data))| logical | numeric | date | string |
|---|---|---|---|
| str | str | str | str |
| "TRUE" | "1" | "2021-01-15" | "abc" |
| "false" | "4.5" | "2021-02-15" | "def" |
| "T" | "Inf" | "2021-02-16" | "ghi" |
This heuristic works well if you have a clean dataset, but in real life, you’ll encounter a variety of challenges that require special handling.
5.2.2 Missing values, column types, and problems
The most common way column detection fails is that a column contains
unexpected values, and you get a string column instead of a more
specific type. One of the most common causes is a missing value recorded
using something other than the None that Polars
expects.
Take this simple 1-column CSV file as an example:
simple_csv = """x
10
.
20
30"""
# Read without specifying null values
pl.read_csv(io.StringIO(simple_csv))| x |
|---|
| str |
| "10" |
| "." |
| "20" |
| "30" |
If we read it without any additional arguments, x
becomes a string column. In this very small example, you can easily see
the missing value .. But what if you have thousands of rows
with only a few missing values represented by .s scattered
throughout?
In Polars, you can specify the column type and tell it what values should be treated as null:
pl.read_csv(
io.StringIO(simple_csv),
schema_overrides={"x": pl.Int64},
null_values=["."],
)| x |
|---|
| i64 |
| 10 |
| null |
| 20 |
| 30 |
5.2.3 Column types
Polars provides many data types that you can specify:
pl.Boolean: For logical values (True/False)pl.Int8,pl.Int16,pl.Int32,pl.Int64: For integers of different sizespl.UInt8,pl.UInt16,pl.UInt32,pl.UInt64: For unsigned integers (positive only)pl.Float32,pl.Float64: For floating-point numberspl.Decimal: For precise decimal arithmeticpl.Utf8: For string datapl.Categorical: For categorical (factor) datapl.Date,pl.Time,pl.Datetime: For date and time datapl.Object: For Python objects that don’t fit the other typespl.Binary: For binary datapl.Struct: For nested data structures
You can specify column types in two ways:
- Using the
schemaorschema_overridesparameter when reading data - Using
cast()to convert columns after reading
# Specify types when reading
csv_data = """x,y,z
1,2,3"""
df = pl.read_csv(
io.StringIO(csv_data),
schema_overrides={
"x": pl.Int32,
"y": pl.Float64,
"z": pl.Utf8
}
)
print(df.schema)
# Or convert after reading
df = pl.read_csv(io.StringIO(csv_data))
df = df.with_columns([
pl.col("x").cast(pl.Int32),
pl.col("y").cast(pl.Float64),
pl.col("z").cast(pl.Utf8)
])
print(df.schema)Schema({'x': Int32, 'y': Float64, 'z': String})
Schema({'x': Int32, 'y': Float64, 'z': String})You can also specify a schema for all columns at once:
schema = {"x": pl.Int32, "y": pl.Float64, "z": pl.Utf8}
df = pl.read_csv(io.StringIO(csv_data), schema=schema)
print(df.schema)Schema({'x': Int32, 'y': Float64, 'z': String})If you want to select only specific columns when reading a file, you
can use the columns parameter:
df = pl.read_csv(io.StringIO(csv_data), columns=["x"])
print(df)shape: (1, 1)
┌─────┐
│ x │
│ --- │
│ i64 │
╞═════╡
│ 1 │
└─────┘5.3 Reading data from multiple files
Sometimes your data is split across multiple files instead of being
contained in a single file. For example, you might have sales data for
multiple months, with each month’s data in a separate file:
01-sales.csv for January, 02-sales.csv for
February, and 03-sales.csv for March.
With Polars, you can read these files one by one and then concatenate them:
# List of sales files
sales_files = [
"data/01-sales.csv",
"data/02-sales.csv",
"data/03-sales.csv"
]
# Read and concatenate via list comprehension
dfs = [pl.read_csv(file).with_columns(pl.lit(os.path.basename(file)).alias("file"))
for file in sales_files]
sales = pl.concat(dfs)This method uses something called List Comprehension, which you can learn more about here. We won’t need these techniques for this class, but you should be aware of them.
You can download these files from the URLs manually, or read them directly with Polars:
sales_files = [
"https://pos.it/r4ds-01-sales",
"https://pos.it/r4ds-02-sales",
"https://pos.it/r4ds-03-sales"
]
dfs = [pl.read_csv(file).with_columns(pl.lit(file).alias("file"))
for file in sales_files]
sales = pl.concat(dfs)If you have many files you want to read in, you can use the
glob module to find the files by matching a pattern:
import glob
sales_files = glob.glob("data/*-sales.csv")
sales_files5.4 Writing to a file
Polars provides several functions for writing data to disk. The most
common are write_csv() and write_parquet().
The most important arguments to these functions are the
DataFrame to save and the file path where you want to save
it.
students.write_csv("students.csv")Note that the variable type information you set up is lost when you save to CSV because you’re starting over with reading from a plain text file. This makes CSVs somewhat unreliable for caching interim results. You need to recreate the column specification every time you load in.
There are two better alternative is Parquet files. These maintain the column types and are generally faster and more space-efficient.
students.write_parquet("students.parquet")
parquet_students = pl.read_parquet("students.parquet")Parquet is becoming more common in use with data sciences, but they have not caught on with more business based analytical tools like Excel, Tableau, & Power BI, so CSVs still reign supreme.
5.5 Data entry
Sometimes you’ll need to assemble a DataFrame “by hand”
with a little data entry in your Python script. Polars provides a simple
way to create data frames from dictionaries where each key is a column
name and each value is a list of values:
df = pl.DataFrame({
"x": [1, 2, 5],
"y": ["h", "m", "g"],
"z": [0.08, 0.83, 0.60]
})
df| x | y | z |
|---|---|---|
| i64 | str | f64 |
| 1 | "h" | 0.08 |
| 2 | "m" | 0.83 |
| 5 | "g" | 0.6 |
You can also create a data frame from a list of dictionaries, where each dictionary represents a row:
df = pl.DataFrame([
{"x": 1, "y": "h", "z": 0.08},
{"x": 2, "y": "m", "z": 0.83},
{"x": 5, "y": "g", "z": 0.60}
])
df| x | y | z |
|---|---|---|
| i64 | str | f64 |
| 1 | "h" | 0.08 |
| 2 | "m" | 0.83 |
| 5 | "g" | 0.6 |
5.6 Summary
In this section, you’ve learned how to load CSV files with Polars’
pl.read_csv() and how to do your own data entry by creating
DataFrames from dictionaries and lists. You’ve learned how CSV files
work, some of the problems you might encounter, and how to overcome
them.
You’ll revisit data import in various formats throughout your Python data science journey, including Excel files, databases, Parquet files, JSON, and data from websites.
Polars is an excellent choice for data handling, as it’s designed to take advantage of modern hardware through parallel processing and memory efficiency. As you become more familiar with Python data science tools, you’ll appreciate Polars’ performance benefits and elegant API design.
6 Exercises
Does it matter what order you used
DataFrame.filter()andDataFrame.sort()if you’re using both? Why/why not? Think about the results and how much work the functions would have to do.What happens if you specify the name of the same variable multiple times in a
DataFrame.select()call?Using the
penguinsdataset, make a scatter plot ofbill_depth_mmvs.bill_length_mm. That is, make a scatter plot withbill_depth_mmon the y-axis andbill_length_mmon the x-axis. Describe the relationship between these two variables.From the
flightsdataset, comparedep_time,sched_dep_time, anddep_delay. How would you expect those three numbers to be related?In a single pipeline for each condition, find all
flightsthat meet the condition:- Had an arrival delay of two or more hours
- Flew to Houston (IAH or HOU)
- Were operated by United, American, or Delta
- Departed in summer (July, August, and September)
- Arrived more than two hours late but didn’t leave late
- Were delayed by at least an hour, but made up over 30 minutes in flight
Find the flights that are most delayed upon departure from each destination.
How do delays vary over the course of the day? Illustrate your answer with a plot.